U-M and multinational coalition of scientists reveal ribosome-RNA polymerase structural link to protein synthesis initiation in bacteria
29 November 2024
An international consortium of researchers from the University of Michigan, the Université de Strasbourg, and the Technische Universität Berlin uncover important players in the mechanistics and the machinery involved in the bacterial ribosome-mRNA translation initiation process through an intercollegiate collaborative new study.
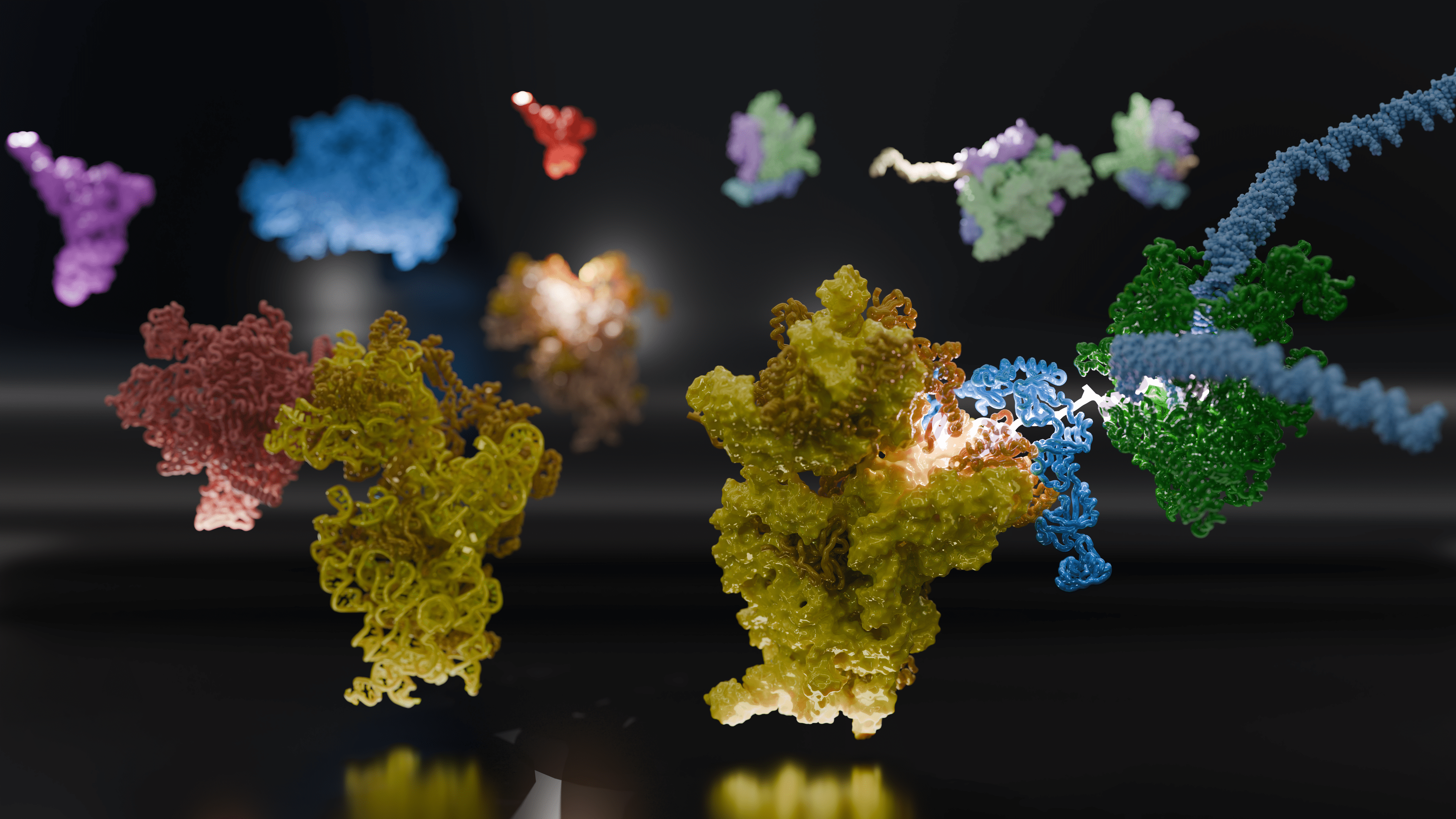
Adrien Chauvier, Ph.D., is a Senior Scientist in the Nils Walter Lab at the University of Michigan, and one of four lead co-authors on a new paper published today in Science. The article, “Molecular basis of mRNA delivery to the bacterial ribosome,” details remarkable findings from a study that sought to visualize the early stages of ribosome recruitment to the nascent messenger RNA (mRNA) as it is transcribed by RNA polymerase (RNAP) and close the gap on a poorly understood aspect of the coupling of these two sequential steps needed to first transcribe mRNA from the genome, then translate it into proteins in the bacterial cell.

Senior Scientist, Nils Walter Lab,
University of Michigan
The research endeavor was led by Albert Weixlbaumer, Ph.D., from the Institut de génétique et de biologie moléculaire et cellulaire in France, Huma Rahil, a Ph.D. student, and Michael Webster, Ph.D., a then-postdoctoral fellow in his lab, from The John Innes Centre in the United Kingdom.
“We wanted to find out how the coupling of RNAP and the ribosome is established in the first place,” Weixlbaumer said. “Using purified components, we re-assembled the complex — ten-billionth of a meter in diameter. We saw them in action using cryo-electron microscopy (cryo-EM) and interpreted what they were doing. We then needed to see if the behavior of our purified components could be recapitulated in different experimental systems.”

©Alexandre Darmon/Art in Research pour la Fondation Bettencourt Schueller
Weixlbaumer and Webster enlisted the expertise of Chauvier and Center for RNA Biomedicine Co-Director Nils Walter, Ph.D., from the University of Michigan in the USA; Juri Rappsilber, Ph.D., from Technische Universität Berlin; and a coalition of scientists from France, the US, Germany, and the United Kingdom.
Walter and Chauvier previously investigated how structural RNAs in bacteria modulate protein production through transcription-translation coupling. Their results were published in The Proceedings of the National Academy of Sciences (PNAS) in 2021.
“Albert was very interested in our work, and wrote a commentary on our paper,” Chauvier said. “It was humbling. I had followed his work for years and was thrilled when he later reached out. His lab analyzes the structure of the ribosome-RNAP complex ‘flash-frozen’ in time, and we look at the dynamic kinetics of that complex ‘in motion.’ It’s exciting how things like this come together. It’s what I love about science.”
Using bacteria as a model system, Weixlbaumer, Chauvier, and the team embarked on a joint project to shed light on this intricate interplay and to apply what they learned in more complex cellular systems. They posed several questions: exactly when, where, and how does the ribosome interact and bind to the newly synthesized mRNA, and what does that look like?
Weixlbaumer explained, “Understanding how the ribosome captures or ‘recruits’ the mRNA is a prerequisite for everything that comes after, such as understanding how it can even begin to interpret the information encoded in the mRNA. It’s like a book. Your task is to read and interpret a book, but you don’t know where to get the book from. How is the book delivered to the reader?”
The researchers discovered that the RNAP transcribing the mRNA deploys two different anchors to rope in the ribosome and ensure a solid footing and start of protein synthesis. This is similar to a foreperson at a construction site overseeing workers installing a complex section of the superstructure, confirming in two redundant ways that all the pieces are fastened securely at critical junctures for maximum stability and functionality.

Image courtesy of Mohammad Afsar and Huma Rahil.
For the study, the team developed a mechanistic framework to show how the various components of the complex work together to bring freshly transcribed mRNAs to the ribosome and act as bridges between transcription and translation.
In more complex human cells, DNA resides in the walled-off nucleus, where RNAP serves as the “interpreter,” breaking down genetic instructions into smaller bites. This dynamo of an enzyme masterfully transcribes, or writes, the DNA into mRNA, representing a specifically selected copy of a small fraction of the genetic code that is moved to the ribosome in the much “roomier” cytoplasm, where it is translated into proteins, the basic building blocks of life.
In prokaryotes, which lack a distinct nucleus and internal membrane “wall”, transcription and translation happen simultaneously and in close proximity to each other, allowing the RNAP and the ribosome to directly coordinate their functions and cooperate with each other.
Bacteria are the best-understood prokaryotes, and because of their simple genetic structure, provided the team with the ideal host to analyze the mechanisms and machinery involved in the ribosome-RNAP coupling during gene expression.
The researchers employed various technologies and methodologies per each lab’s specialty — cryo-EM in Weixlbaumer’s group, and the Berlin group’s in-cell crosslinking mass spectrometry carried out by Andrea Graziadei — to examine the processes involved.
With expertise in biophysics, Chauvier and Walter utilized their advanced single molecule fluorescence microscopes to analyze the kinetics of the structure.
Chauvier explained, “We used single-molecule colocalization between an RNAP paused elongation complex (PEC) and the ribosome — one fluorescent color for the nascent mRNA emerging from RNAP and one for the ribosome — to track the kinetics of association and dissociation between the two machineries.”

They observed that the mRNA emerging from RNAP was bound to the small ribosomal subunit (30S) particularly efficiently when ribosomal protein bS1 was present, which helps the mRNA unfold in preparation for translation inside the ribosome.
The cryo-EM structures of Webster and Weixlbaumer pinpointed an alternative pathway of mRNA delivery to the ribosome, via the tethering of RNA polymerase by the coupling protein factor NusG, or its paralog, or version, RfaH, which thread the mRNA into the mRNA entry channel of the ribosome from the other side of bS1.

John Innes Centre, United Kingdom
Webster said, “We call the series of ordered, controlled, and interconnected events a ‘molecular mechanism,’ to emphasize how it can be thought of like the complex mechanism inside a clock for example. I am always amazed that it is possible to reconstitute such an exquisite and biologically fundamental process in a test tube. It is particularly exciting now to have the opportunity to use powerful imaging techniques to answer questions that researchers have been interested in for several decades.”
Understanding these fundamental processes holds great potential for developing new antibiotics that target these specific pathways in bacterial protein synthesis. Traditionally, antibiotics have targeted the ribosome or RNAP, but bacteria often find a way to evolve, and mutate to create some resistance to those antibiotics. Armed with their new knowledge, the team hopes to outwit bacteria by cutting off multiple pathways.
“We know there is an interaction between the RNAP, the ribosome, transcription factors, proteins, and mRNA,” Chauvier said. “We could target this interface, specifically between the RNAP, ribosome, and mRNA, with a compound that interferes with the recruitment or the stability of the complex.”
Although Weixlbaumer initiated the study with no particular application in mind, he underscored the crucial role fundamental research plays in tackling scientific challenges. He said, “Our work is curiosity-driven, understanding very fundamental processes. It’s like repairing a car, If you don’t comprehend how it works first, you can’t even begin to think about how to fix it.”
Having successfully visualized the very first stage in establishing the coupling between RNAP and the ribosome, the team looks forward to further collaboration to find out how the complex needs to rearrange to become fully functional. Weixlbaumer added, “The book is delivered. How do we open to page one and start reading?”
“This work demonstrates the power of interdisciplinary research carried out across continents and oceans,” said Walter.

Francis S. Collins Collegiate Professor of Chemistry, Biophysics and Biological Chemistry, Professor of Chemistry, Professor of Biophysics, College of
Literature, Science,
and the Arts, University of Michigan
Read the Michigan News announcement – Office of the Vice President for Communications, University of Michigan.
The work was supported by grants from the Agence Nationale de la Recherche (France); Zentrales Innovationsprogramm Mittelstand (ZIM) des Bundesministeriums für Wirtschaft und Klimaschutz (Germany); European Research Council; the National Institutes of Health (United States); and the National Science Foundation.
Collaborative New Study:
M. Webster, A. Chauvier, et. al. Molecular basis of mRNA delivery to the bacterial ribosome, Science, 29 November 2024, http://www.science.org/doi/10.1126/science.ado8476
2021 University of Michigan Study:
S. Chatterjee, A. Chauvier, S.S. Dandpat, I. Artsimovitch, N.G. Walter, A translational riboswitch coordinates nascent transcription–translation coupling, Proc. Natl. Acad. Sci. U.S.A. 118 (16) e2023426118, https://doi.org/10.1073/pnas.2023426118 (2021).
Read the full story of the 2021 study by Morgan Sherburne, Michigan News.