U-M Faculty Members
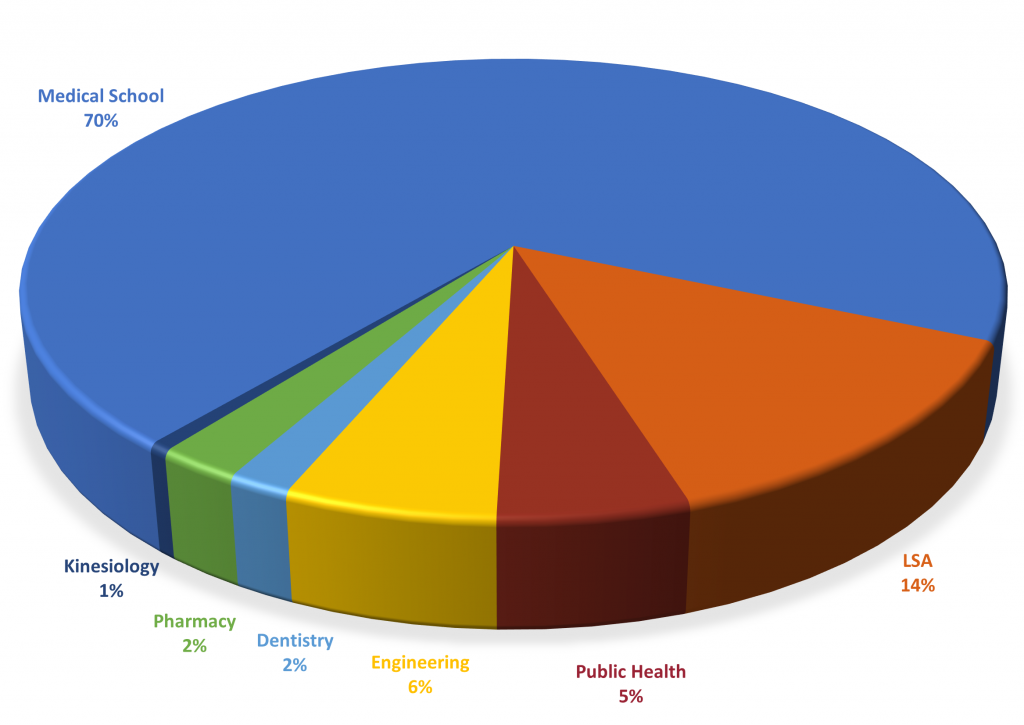
Over 170 faculty have joined the Center for RNA Biomedicine. They belong to seven schools and colleges across the University of Michigan.
This large membership demonstrates the central role played by RNAs as well as the need for collaborations across disciplines to study the complexity of RNA biological processes. One of the goals of the center is to foster and support such collaborations and synergies.
All University of Michigan faculty who study RNAs are welcome to join this community. Learn more about our membership and its benefits here.
| First Name | Last Name | Unit/College | Department | Research Description |
|---|---|---|---|---|
| Carlos | Aguilar | Engineering | Biomedical Engineering | The long-term goal of the NOBEL is to make breakthroughs in medicine and biology that instill hope and inspire others. To accomplish this feat, we develop, optimize and apply innovative technologies such as integrative genomic assays and high-throughput sequencing, micro/nanofabricated devices, genome editing and computational modeling to our primary area of focus, which is skeletal muscle. Lab Website Michigan Research Experts Profile Featured Scientist |
| Huda | Akil | Medical School | Molecular & Behav Neurosci Inst | Research in the Akil laboratory is focused on understanding the neurobiology of emotions, including pain, anxiety, depression and substance abuse. Early on, our research focused on the role of the endorphins and their receptors in pain and stress responsiveness. We provided the first physiological evidence for a role of endogenous opioids in the brain; and showed that endorphins are activated by stress and cause pain inhibition, a phenomenon we termed Stress-Induced Analgesia. We defined how the posttranslational processing of opioid precursors is modulated by stress, and demonstrated the coordinate actions of the neuropeptide products on behavior. Website Michigan Research Experts Profile |
| Benjamin | Allen | Medical School | Cell & Dev Biology | Research in the Allen Lab is broadly focused on understanding the mechanisms of growth factor and morphogen signaling in development and disease. Specifically, we study the regulation of Hedgehog (Hh) signaling during vertebrate embryogenesis using a wide range of approaches, including mouse developmental genetics, chick in ovo electroporation, biochemistry, and cell biology. The long-term goal of our research is to use insights gained from the study of Hh signaling in normal development in the treatment of a broad spectrum of developmental diseases and childhood and adult cancers. Website Michigan Research Experts Profile |
| Joshi | Alumkal | Medical School | Int Med | The Alumkal Laboratory’s main emphasis is to understand how genomic and epigenomic changes contribute to lethal prostate cancer progression. We use biochemical, genomic, and epigenomic approaches in cellular models to clarify mechanisms by which key transcriptional regulators function. The most important goals of our studies are to identify therapies that effectively target and interdict lethal prostate cancer progression and to conduct innovative clinical trials designed to validate the molecular and clinical effectiveness of novel therapeutic agents. Lab Website Michigan Research Experts Profile Featured scientist Video |
| Anthony | Antonellis | Medical School | Human Genetics | Our group is broadly interested in how human genetic variation affects gene and genome function. One area of research is grounded in the observation that mutations in aminoacyl-tRNA synthetases cause myriad disease phenotypes including dominant axonal neuropathy and recessive syndromes that include neurodevelopmental defects. A second area of research aims to study how gene regulation—via cis-acting transcriptional regulatory elements including enhancers and promoters—plays a role in the development of myelinating cells of the peripheral and central nervous systems. We are always looking for outstanding students and post-doctoral fellows to advance each of these areas. Lab Website Michigan Research Experts Profile Featured scientist |
| Brian | Athey | Medical School | Comp Med and Bioinformatics | One main area of research is the application of high resolution fluorescence optical microscopy coupled with high-throughput analysis, 3D imaging and machine learning to explore the chromatin structure and nuclear architecture of cells. This research emphasizes the convergence between 3D structural predictions and 3D structural measurements with microscopy, to provide insight into the transcriptional architecture of the interphase nucleus. The next area of research centers on the exploration of the pharmacoepigenome in psychiatry, neurology, anesthesia and addiction medicine. This research employs high-throughput 4D microscopic imaging of enhancers, promoters and chromatin features, using fluorescence in situ hybridization (FISH). These methods are coupled with Hi-C chromatin conformation capture, chromatin state annotation, localization in postmortem human brain tissue and induced neuronal pluripotent stem cells, and machine learning for identification of regulatory variants, to provide insight into the genetic and epigenetic mechanisms of inter-individual and inter-cohort differences in psychotropic drug response. Lab Website Michigan Research Experts Profile Google Scholar |
| Sara Jo | Aton | LSA | Molecular, Cellular, and Developmental Biology | Brain states such as sleep and wakefulness have a profound impact on mRNA transcription and translation. Research in the Aton lab characterizes how sleep contributes to transcription and translation of mRNAs in different cell types of the central nervous system, how these processes affect learning and memory consolidation, and how they are affected by genetic variants associated with disorders of neurodevelopment. Lab Website Michigan Research Experts Profile Google Scholar Featured scientist |
| Ryan | Bailey | LSA | Chemistry | The Bailey group develops and deploys multiplexed analytical tools to quantitate miRNA, mRNA, and lncRNA signatures of health and disease. They also are interested in probing mechanisms of epigenetic regulation of gene expression. Lab Website Google Scholar |
| James | Bardwell | LSA | Molecular, Cellular, and Developmental Biology | Our lab focuses on chaperone biology, we are currently investigating our surprising finding that RNA may play a significant role in protein folding in vivo. Nucleic Acids Research, 2016 Jun 2;44(10):4835-4845 Publication Lab Website Michigan Research Experts Profile Google Scholar |
| Sami | Barmada | Medical School | Neurology | RNA decay is critical for the maintenance of neuronal health and function. Our laboratory identified fundamental RNA decay abnormalities in the neurodegenerative diseases ALS and FTD, and is now focused on determining the etiology and consequences of these changes for neuronal survival and disease progression. Lab Website View Michigan Research Experts Profile Featured scientist |
| Scott | Barolo | Medical School | Cell & Dev Biology | Transcriptional regulation of gene expression; cis-regulatory elements (enhancers) Faculty Profile Michigan Research Experts Profile Google Scholar |
| Stuart | Batterman | Public Health | Environmental Health Sciences | My current research addresses a range of topics in occupational, indoor and environmental settings that include: Exposure assessment (especially for volatile organic compounds or VOCs); emerging contaminants in occupational and environmental settings (e.g., brominated flame retardants); biological monitoring; air quality monitoring; indoor air quality (e.g., assessment and management); air pollution control engineering (e.g., vapor and particle air filtration); and environmental epidemiology. Website Miichigan Research Experts Profile Google Scholar |
| Allison | Billi | Medical School | Dermatology | Dr. Billi’s research explores the biological mechanisms that lead to female sex bias in autoimmune diseases such as systemic lupus erythematosus (SLE). Dr. Billi has shown that overexpression of the transcription cofactor VGLL3 in the skin of mice causes a severe lupus-like rash and systemic autoimmune disease that mimics SLE in humans. In addition, she has shown that excess epidermal VGLL3 drives a program of proinflammatory gene expression that overlaps in both female skin and cutaneous lupus. Importantly, these findings strongly suggest VGLL3 serves a central role in causing lupus in women and that skin inflammation can precipitate systemic autoimmune disease. Website Faculty Spotlight Google Scholar |
| Markus | Bitzer | Medical School | Int Med Nephrology | My research is focus on understanding the functions of non-coding RNA in kidney disease and aging. We have established an active biobank of currently over 100 human kidney tissue samples, which we are using to integrate transcriptomic profiles generated by microarray and RNA-sequencing, with quantitative morphometric (morphomic) data. Findings are validated in tissue culture and animal models of kidney injury and aging using genetic model systems as well as RNA-inhibitors and mimics. Website Michigan Research Experts Profile Google Scholar |
| Alan | Boyle | Medical School | Comp Med and Bioinformatics | We use modern genomics techniques and high-throughput experiments to explore biological systems. We can leverage computational tools to help answer biological problems that were previously intractable. We aim to combine computational approaches with high-throughput biological assays to better understand the whole human transcriptional regulatory system. Lab Website Google Scholar Featured scientist |
| Charles | Brooks | LSA | Chemistry | Our group studies RNA structure, dynamics and folding using both all atom and coarse grained models. Current interests involve the assembly of the ribosome and it’s functional dynamics, the folding of complex RNA junction topologies, pH effects in RNA catalysis and protein-RNA/DNA interactions in translation and transcription. Lab Website Google Scholar Featured scientist |
| Charles | Burant | Medical School | Internal Medicine | Clinical interests are in the care of patients with obesity, type 2 diabetes, and related conditions. Burant’s research program integrates molecular phenotyping (genomics, transcriptomics, proteomics, and transcriptomics) with dietary, clinical, and biobehavioral phenotypes to understand the development of obesity, insulin resistance, and diabetes in both people and in animal models. Director of the Taubman Institute, which is dedicated to fundamental discovery, and to advancing our knowledge of the causes, treatment, and prevention of diseases and conditions that affect mankind. Website |
| Margit | Burmeister | Medical School | Molecular & Behav Neurosci Inst | In identifying genetic variants associated with Mendelian (e.g. Ataxia) and complex (e.g depression and addiction), often the variant in question is not protein coding. We have found, for example in 2010, that a 5’UTR variant increased mRNA expression 2-3 fold, resulting in deafness. We have published on miRNA expression differences possibly related to Bipolar disorder, and are involved in several projects, both related to ataxia and to addiction, in which we suspect that mRNA regulation plays a role. Website Michigan Research Experts Profile |
| Mark | Burns | Engineering | Chemical Engineering | Currently, I have some work in sensing RNA-containing pathogens (e.g., influenza), and I am considering expanding my work in this area. Lab Website Michigan Research Experts Profile Google Scholar |
| Laura | Buttitta | LSA | Molecular, Cellular, and Developmental Biology | In the Buttitta Lab we are studying how gene expression at the mRNA level changes when cells enter quiescent or non-dividing states. We are also interested in the transcription factors and chromatin remodelers that may be influenced by cell cycle regulators to mediate changes in mRNA when cells enter quiescence. Lab Website View Michigan Research Experts Profile |
| Dawen | Cai | Medical School | Cell & Dev Biology | Single cell RNA profiling to identify neuronal stem/mature cell types and markers. Lab Website Michigan Research Experts ProfileGoogle ScholarFeatured scientist |
| Sally Ann | Camper | Medical School | Human Genetics | Interested in single cell RNA sequencing for understanding cell differentiation in health and disease Lab Website Michigan Research Experts Profile |
| Maria | Castro | Medical School | Neurosurgery | Glioma genetic models are needed to uncover mechanisms which mediate tumor progression, interplay with the tumor microenvironment (TME) and response to therapeutics. We have generated the first genetically engineered immunocompetent mouse model of low grade glioma(LGG), harboring mIDH1 in combination with ATRX and p53 knock-down. We have generated primary neurospheres (NS) from LGG, which exhibit cancer stem-cell- like properties, and have enabled us to develop a transplantable model of LGG, amenable to testing novel therapies. NS are derived from C57BL/6 mice, thus, enabling examination of the immune TME and the impact of tumor mutations on the immune response. Our goals are to assess the effect of mIDH1 on mRNA-seq, and on global DNA and histone methylation. The mIDH1 model will also be used for chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) to identify promoter/enhancer region specific changes in histone methylation. We are collaborating with Dr. Mats Ljungman who pioneered bromouridine sequencing (BrU-seq), to identify and quantify nascent mRNA and gene transcription profiles. Uncovering epigenetic patterning of histone 3 hypermethylation and cytosine modifications using next generation sequencing (NGS) technologies will contribute to the identification of novel pathways and gene regulatory networks which will provide novel insights into disease biology and uncover novel therapeutic targets. Lab Website Michigan Research Experts Profile Featured scientist |
| Sriram | Chandrasekaran | Engineering | Biomedical Engineering | We are at the Department of Biomedical Engineering in the University of Michigan, Ann Arbor. Our lab develops computer models for drug discovery and understanding cellular metabolism. Our recent research includes designing drug combinations for antibiotic resistant bacteria using AI and discovering a new mechanism of pathogen clearance by killer T-cells. We are also developing new methods to simulate the metabolism of microbes, cancer- and stem-cells. WebsiteGoogle Scholar |
| Matt | Chapman | LSA | Molecular, Cellular, and Developmental Biology | The Chapman lab is interested in the production, turnover and processing of RNA during bacterial biofilm formation. Lab Website |
| Grace | Chen | Medical School | Int Med | I am interested in how certain circulating non-coding small RNAs regulate colorectal cancer progression. Lab Website Michigan Research Experts Profile |
| Arul | Chinnaiyan | Medical School | Pathology | Dr. Chinnaiyan’s research is focused on functional genomic, proteomic and bioinformatics approaches to study cancer for the purposes of understanding cancer biology as well as to discover clinical biomarkers. His group has characterized a number of biomarkers of prostate cancer including AMACR, EZH2, hepsin and sarcosine. AMACR is being used clinically across the country in the assessment of cancer in prostate needle biopsies. The landmark study from Dr. Chinnaiyan’s lab thus far is the discovery of TMPRSS2-ETS gene fusions in prostate cancer. TMPRSS2-ETS gene fusions are specific markers of prostate cancer as well as presumably function as rational targets for this disease. This finding potentially redefines the molecular basis of prostate cancer as well as other common epithelial cancers. Currently efforts are underway to target this gene fusion as well as discover similar gene fusions in other common epithelial tumors such as those derived from the breast, lung, and colon. Website Michigan Research Experts Profile The Michigan Center for Translational Pathology |
| Michael | Cianfrocco | Medical School | Biological Chemistry | Motor protein dependent RNA localization & structural biology using cryo-electron microscopy Lab Website Michigan Research Experts Profile |
| Justin | Colacino | Public Health | Environmental Health Sciences | Our goal is to characterize the environmental susceptibility of normal human stem cell populations, elucidating the etiology of sporadic cancers. Of particular interest are understanding the changes that occur at the epigenomic and transcriptomic level, changes which affect not only gene expression but also how progenitor cells differentiate and divide. Lab Website Michigan Research Experts Profile |
| Kathleen | Collins | Medical School | Int Med Nephrology | We study the ability of HIV to establish a persistent infection in long lived cells. We are interested in novel approaches to detect and eradicate these cells. Website Michigan Research Experts Profile Lab Website |
| Analisa | DiFeo | Medical School | Pathology | My laboratory investigates epithelial ovarian cancer (EOC) biology at the interface between the bench and the clinic in order to generate findings that can directly benefit ovarian cancer patients. In order to accomplish this we focus on four major areas: 1) generation of clinically relevant EOC models, 2) development of novel or re-purposed drugs that can work alone or in conjunction with current treatment options to combat this deadly disease, 3) discovery of potent drivers of drug resistance and recurrence, and 4) identify novel biomarkers for early detection or therapeutic response. We have uncovered that microRNA-181a is frequently overexpressed in recurrent, platinum-resistant HGSOC tumors and correlates with shorter time to recurrence and poor overall survival. Functionally, we found that it can modulate several potent cancer-associated pathways such as TGF-β and Wnt signaling which in turn leads to the induction of epithelial-to-mesenchymal transition (EMT), drug resistance and increase tumor-initiating capacity. Most recently, we have developed a unique method to isolate primary tumor cells based miRNA function and identify drugs that regulate its expression using miRNA biosensors. Through these studies we uncovered that the miR-181a promoter contains a super enhancer which is targeted by Bromodomain inhibitors (BETi). Lab Website Featured scientist Video |
| Dana | Dolinoy | Public Health | Environmental Health Sciences | Our research seeks to advance development of a suite of tools, based on PIWI-interacting RNA (piRNA) to transform precision environmental health and epigenome editing. Faculty Profile Lab Website Michigan Research Experts ProfileGoogle Scholar |
| Gregory | Dressler | Medical School | Pathology | The Dressler lab studies genes and pathways that regulate not only embryonic development and but also contribute to adult disease. Aspect currently being investigated by genetic and biochemical means are the structure and functions of long 5′ untranslated regions of specific mRNAs and how these complex regions impact translation and gene expression of key regulatory proteins. Website Michigan Research Experts Profile |
| Monica | Dus | LSA | Molecular, Cellular, and Developmental Biology | The Dus lab is interesting in understanding how the environment reshapes brain and behavior. To this end we are studying how diet changes the physiology of the brain by altering metabolic-epigenetic pathways at both the transcriptional and post-transcriptional levels, and how this, in turn, affects behavior. Lab Website Michigan Research Experts Profile Featured scientist |
| James | Elder | Medical School | Dermatology | Our laboratory is interested in molecular mechanisms controlling epidermal growth and differentiation, including how this process is linked to host defense and autoimmunity. For this purpose, we utilize cell biology, organ culture, transgenic animals, genetic linkage analysis, and gene expression profiling. Website |
| Eric | Fearon | Medical School | Int Med Nephrology | The Fearon laboratory’s interests include the role of non-coding RNAs regulated by key oncogene and tumor suppressor gene pathways in colorectal cancer development and in invasive and metastatic phenotypes. In addition, the Fearon laboratory is utilizing CRISPR-Cas9 approaches to generate novel genetically engineered mouse models of colorectal cancer as well as the role of collaborative somatic oncogene and tumor suppressor gene defects in tumor initiation and clonal evolution. Website Michigan Research Experts Profile |
| Eva | Feldman | Medical School | Neurology | Amyotrophic lateral sclerosis (ALS) is a progressive degeneration of motor neurons that develops from a complex interaction of genetic, epigenetic, and environmental factors. We are investigating the role of altered RNA and protein metabolism in ALS by focusing on mRNA stability, microRNA dysregulation, RNA exosomes, and microRNA/protein interactions in patient tissues and cellular models. Our study of RNA in ALS will help identify early diagnostic biomarkers and elucidate pathological mechanisms that could point to possible therapeutic targets. Lab Website Michigan Research Experts Profile Featured scientist |
| Lydia | Freddolino | Medical School | Biological Chemistry | Measurement of protein-RNA and RNA-RNA interactions driving microbial regulatory networks. Lab Website Michigan Research Experts Profile Featured scientist |
| Santhi | Ganesh | Medical School | Human Genetics | The Ganesh lab studies transcriptional responses to vascular injury and stress, and cellular intervention targets to ameliorate vascular diseases. Lab Website Michigan Research Experts Profile |
| George | Garcia | Pharmacy | Medicinal Chemistry | In the past, we have studied post-transcriptional modification of tRNA. Currently we are pursuing RNA polymerase as a drug target for tuberculosis. Lab Website Michigan Research Experts Profile |
| Amanda | Garner | Pharmacy | Medicinal Chemistry | The Garner Laboratory is developing novel high-throughput screening assays for the discovery of chemical probes for targeting microRNAs and microRNA-microRNA-binding protein interactions. In addition, we are engaged in the discovery of new microRNA-binding proteins using chemical biology and proteomic approaches. Lab Website Michigan Research Experts Profile Featured scientist |
| Scott | Gitlin | Medical School | Int Med Nephrology | Retroviruses; viral gene expression. Website Michigan Research Experts Profile Markovitz Research Group |
| Thomas | Glover | Medical School | Human Genetics | Our research focuses on the mechanisms and consequences of genome instability and on the molecular biology of human genetic disease. A longstanding interest is the study of chromosome damage following replication stress. This began with the study of chromosome fragile sites and has led to our current focus on copy number variants (CNVs). Website Michigan Research Experts Profile |
| Daniel | Goldman | Medical School | Neuroscience | Molecular mechanisms underlying retina and optic nerve regeneration in fish and mammals. Regarding RNA, our research studies transcriptional and posttranscriptional mechanisms contributing to regeneration, including microRNAs and other non-coding RNAs. Website Michigan Research Experts Profile |
| Stephen | Goutman | Medical School | Neurology | Dr. Goutman completed his medical degree from the University of Chicago’s Pritzker School of Medicine, and completed his neurology residency and a Fellowship in Neuromuscular Disease at the Cleveland Clinic Hospital in Cleveland, Ohio. His clinical and research interests focus on amyotrophic lateral sclerosis (ALS). Website Michigan Research Experts Profile |
| Yuanfang | Guan | Medical School | Comp Med & Bioinformatics | Functional genomic data, such as RNA-seq, microarray, protein-protein interactions, are growing exponentially these days. The research in Guan Lab focuses on developing novel and high-accuracy algorithms that integrate these data for predicting gene functions and networks. Lab Website Michigan Research Experts Profile |
| Johann | Gudjonsson | Medical School | Dermatology | Gudjonsson’s lab focuses on analysis of coding and non-coding RNA transcription in cells and tissues, and addressing their role in normal and diseased state, with a particular focus on skin. Lab Website Michigan Research Experts Profile |
| Gary | Hammer | Medical School | Int Med Nephrology | Dr. Hammer’s research focuses on the molecular underpinnings of adrenocortical growth in development and cancer. His laboratory’s goals are to characterize the adrenocortical stem/ progenitor cell population and elucidate how altered regulation of these cells contributes to adrenocortical disease, namely hypoplasias, dysplasias and cancer. Lab Website Michigan Research Experts Profile |
| Sue | Hammoud | Medical School | Human Genetics | Single Cell RNAseq Website Michigan Research Experts Profile |
| Phyllis | Hanson | Medical School | Biological Chemistry | Broadly interested in the relationship between mRNA and membranes, both within the cell and outside in extracellular vesicles. Website Michigan Research Experts Profile |
| Michelle | Hastings | Medical School | Pharmacology | Research focuses on understanding genetic basis of disease and discovering new therapeutics that modulate the process of pre-mRNA splicing to alter gene expression. Website Michigan Research Experts Profile |
| Alfred | Hero | Engineering | COE EECS – ECE Division | The Hero lab works on translational research that develops algorithms and models for personalized precision medicine that combine data from very different platforms for predicting disease outcomes and contagiousness. Data that we are working with include synchronized genomic modalities such as HiC, RNAseq and ChipSeq, imaging data including FISH and fMRI. Website Michigan Research Experts Profile |
| Gerry | Higgins | Medical School | Comp Med and Bioinformatics | My current research focuses on the regulation of gene expression and the pharmaco-epigenome (e.g. noncoding regulatory variants and elements). My published research has included studies of the mechanisms of alternative splicing of CNS genes. I also teach in the RNA-Seq course (BIoINF 545; BIOSTAT 646) Website Michigan Research Experts Profile |
| Zhonggang | Hou | Medical School | Biological Chemistry | New CRISPR tools for RNA guided genome engineering Website Michigan Research Experts Profile Featured scientist |
| Lori | Isom | Medical School | Pharmacology | We are developing antisense oligonucleotide therapeutic agents to treat developmental and epileptic encephalopathy. Lab Website Michigan Research Experts Profile |
| Shigeki | Iwase | Medical School | Human Genetics | Our group studies histone methylation-mediated mechanisms underlying mRNA production, i.e. transcription, in the brain. Given that many histone methyl regulators are mutated in cognitive disorders, our research may yield important insights into how histone methylation is involved in normal and pathological brain development and function. Lab Website Michigan Research Experts Profile Featured scientist |
| Matthew | Iyer | Medical School | Pathology & Surgery | I am an early-stage surgeon scientist at the University of Michigan dedicated to improving cancer care through the innovation of RNA liquid biopsy assays. My career vision arose from predoctoral and postdoctoral work in the laboratory of Arul Chinnaiyan, M.D., Ph.D. (2009-2016), where I developed RNA-Seq analysis methods to investigate the uncharted regions of the human genome, discovering a multitude of new genes and isoforms with potential applications in cancer characterization. Through this work, I saw opportunities to 1) further expand our knowledge of the cancer transcriptome by integrating extensive molecular data, and 2) translate this new knowledge into improving molecular diagnostic assays. I then devoted myself to surgical residency and fellowship training (2016-2023), becoming proficient in the care of cancer patients and their families. My current research at the University of Michigan aims to improve bioinformatics methods for analyzing bulk, single-cell, and spatial transcriptomic data as well as conduct clinical studies of novel molecular sequencing assays that could inform cancer care. Website |
| Ursula | Jakob | LSA | Molecular, Cellular, and Developmental Biology | The Jakob Lab does research on the biochemical aspects of oxidative stress in various organisms. Lab Website Michigan Research Experts Profile |
| Paul | Jenkins | Medical School | Pharmacology | The Jenkins’s lab is interested in the splicing of neuronal genes and how dysregulation of this process underlies neuropsychiatric disorders. Lab Website Michigan Research Experts Profile |
| Hui | Jiang | Public Health | Biostatistics | My research interest is in developing statistical and computational methods to analyze RNA-Seq data for the quantification of gene and isoform expression and the detection of differentially expressed or spliced genes. Website |
| Catherine | Kaczorowski | Medical School | Neurology | Targeting mRNA for treatment of Alzheimer’s and other neurodegenerative diseases. Lab Website Michigan Research Experts Profile |
| Sundeep | Kalantry | Medical School | Human Genetics | Our research aims to define how epigenetic regulation occurs in mammals. We use X-chromosome inactivation, which results in transcriptional silencing of most genes along one of the two X-chromosomes in female mammals, to gain insights into epigenetic processes, including through chromatin modifications and long-non coding RNAs. Lab Website Michigan Research Experts Profile Featured scientist |
| Hyun Min | Kang | Public Health | Biostatistics | Dr. Kang’s research focuses on developing robust, scalable, and practical methods and tools to analyze large-scale genomic data to help understand the etiology of complex traits. Website Michigan Research Experts Profile |
| Sarah | Kargbo-Hill | LSA | Molecular, Cellular and Developmental Biology | My research focuses on defects in RNA metabolism that occur in neurodegenerative disease. In particular, I focus on the RNA-binding protein TDP-43, which is associated with neurodegenerative disorders including ALS, FTD, and Alzheimer’s disease. I am interested in the role of TDP-43 as a regulator of cryptic splicing, both in understand how these events are regulated in different cells types the consequences of cryptic splicing events on downstream pathways in neurons. I use human iPSC-derived neurons to examine these questions in a human neuron genetic background. Lab Website |
| Sarah | Keane | LSA | Biophysics, Chemistry | Structure and mechanism on noncoding RNAs Lab Website Michigan Research Experts Profile Featured scientist |
| Evan | Keller | Medical School | Urology | Explore single cell transcriptomes through single cell RNASeq. Primarily focused on cancer. Website Michigan Research Experts Profile Lab Website |
| Tom | Kerppola | Medical School | Biological Chemistry | My laboratory investigates the mechanisms whereby transcription factor complexes regulate gene expression Lab Website Michigan Research Experts Profile |
| Jeffrey | Kidd | Medical School | Human Genetics | We study determinants of viral latency and reactivation as well as the contribution retrotransposons to disease, phenotypic diversity, and genome evolution. Lab Website Michigan Research Experts Profile |
| Anthony | King | Medical School | Psychiatry | Gene expression in humans with PTSD Lab Website Michigan Research Experts Profile |
| Jacob | Kitzman | Medical School | Human Genetics | The Kitzman lab develops new experimental and computational approaches to dissect the functional impacts of genetic variants. We are applying these to several clinically-relevant genes and non-coding elements, with the goal of comprehensively measuring the effects of all possible variants at each locus, from their transcription and splicing to the activity of the resulting encoded RNAs and proteins. Website Michigan Research Experts Profile |
| Markos | Koutmos | LSA | Chemistry | RNA processing, RNA modifications Website Michigan Research Experts Profile Featured scientist Lab Website |
| Kristin | Koutmou | LSA | Chemistry | The Koutmou lab studies the essential molecular machine responsible for translating the nucleic acid code into protein, the ribosome. We use a combination of in vitro biochemistry (eg. reconstituted translation system) and genome wide tools (eg. ribosome profiling) to investigate the molecular level details of situations in which translation is dysfunctional. Website Lab Website Featured scientist |
| Matthias | Kretzler | Medical School | Int Med Nephrology | The overarching goal of Dr. Kretzler’s research is to define chronic kidney disease in mechanistic terms and use this knowledge for targeted therapeutic interventions. To reach this goal he and his team have developed a translational research pipeline centered on integrated systems biology analysis of renal disease. Website Michigan Research Experts Profile |
| Chandan | Kumar-Sinha | Medical School | Pathology | Integrative functional-genomics of cancers using high-throughput genome and transcriptome analyses to develop cancer biomarkers and personalized therapeutic options. Website Google Scholar |
| Steve | Kunkel | Medical School | Pathology | Research directed at understanding the expression and regulation of inflammatory mediators and their subsequent biological activities represents major investigative directions of my laboratory. A general theme of the laboratory is an assessment of the mechanistic involvement of cytokine and chemokine cascades that direct the initiation and maintenance of the immune response. Website Michigan Research Experts Profile |
| Kenneth | Kwan | Medical School | Molecular & Behavioral Neurosi Inst | Trafficking and local translation in neurons. The Kwan laboratory at the University of Michigan studies the development, evolution, and dysfunction of neural circuits in the cerebral cortex. Website Michigan Research Experts Profile Lab Website |
| Joerg | Lahann | Engineering | Chemical Engineering | At Lahann lab, we are dedicated to addressing the critical challenges in the field of biotechnology and drug/gene delivery as well as advancing the current knowledge in chemical vapor deposition for engineered surfaces. We are an interdisciplinary research group developing novel and exciting technologies. Lab Website |
| Adam | Lauring | Medical School | Microbiology & Immunology | Adam Lauring’s laboratory studies the population genetics and evolutionary dynamics of RNA viruses, which replicate with extremely low fidelity. Core projects include understanding the molecular determinants of RNA virus mutation rates and the impact of mutations on viral fitness. Lab Website Michigan Research Experts Profile |
| Cheng-Yu | Lee | Medical School | Cell & Dev Biology | We study the mechanisms of neural stem cell self-renewal and differentiation, which have implications for neurological disorders and cancer. Interested in RNA decay and translational control. Lab Website Michigan Research Experts Profile |
| Jun Hee | Lee | Medical School | Molecular & Integrative Physiology | My research indeed focuses on RNA metabolism, stress granules, and single-cell/spatial transcriptomics. Lab Website Michigan Research Experts Profile Featured scientist |
| Jiahe | Li | Engineering | Biomedical Engineering | We address the knowledge gap in understanding the roles played by host-derived extracellular sRNAs in host-microbiota interactions. There is a lack of enabling methodologies to unlock the functions of host sRNAs largely due to three limitations. First, native sRNAs are highly susceptible to degradation. Second, it is difficult to distinguish the origins of short sRNA fragments between host and bacteria. Third, host sRNAs are present inside bacteria at very low levels. Our long-term goal is to achieve in-depth understanding of host-derived sRNAs as a new class of defense molecules targeting opportunistic pathogens, and, furthermore, to repurpose host sRNAs as potential antimicrobial agents. Website |
| Jun | Li | Medical School | Human Genetics | My group has a long-standing interest in classification and unsupervised class discovery, especially for high-dimensional genomics data. I am co-directing the proposed Michigan Center for Single-Cell Genomic Data Analytics, focusing on single-cell RNAseq data and currently including studies of cancer evolution, organ development, gene-environment interaction, and regulation of RNA splicing. Lab Website Michigan Research Experts Profile |
| Yongqing | Li | Medical School | Surgery | Dr. Li has focused his study on pharmacological treatment of sepsis, traumatic hemorrhagic shock, traumatic brain injury (TBI), with emphasis on histone modifying enzymes, including histone deacetylase (HDAC) and peptidyl-arginine deiminase (PAD). His team developed a novel antibody against citrullinated histone H3 (CitH3), which is significantly different from commercially available one. Targeting CitH3 for early diagnosis/prognosis and treatment of sepsis has captured researchers’ attention in both academia and industry. These findings are being translated into the clinic now. Website Michigan Research Experts Profile |
| Jiandie | Lin | Medical School | Cell & Dev Biology | My group studies how long noncoding RNAs (lncRNAs) regulate metabolic gene programs in adipose tissues and the liver. We are interested in dissecting the role of lncRNAs in the control of energy metabolism in physiological and disease states. Lab Website Michigan Research Experts Profile |
| Jie | Liu | Medical School | Comp Med and Bioinformatics | Machine learning, chromatin organization, alternative splicing Lab Website Michigan Research Experts Profile |
| Mats | Ljungman | Medical School | Radiation Oncology | My lab is studying mechanisms of transcriptional and post-transcriptional regulation of gene expression in normal physiology and in diseases such as cancer and ALS. We have developed nascent RNA Bru-seq that we are happy to help other labs use. Lab Website View Michigan Research Experts Profile |
| Pedro | Lowenstein | Medical School | Neurosurgery | Glioma stem cells exist in two states: as stem cells proper, and as brain tumor cells. We are currently working to understand the molecular underpinnings of state change in glioma tumors stem cells. This involves many studies that use RNA-seq, Bru-seq (with Mats Ljungman), and Linc-seq. Studies on self-organization in brain tumors and mechanisms of tumor growth (both in rodents and in humans), and the response of glioma tumors to immune attack, also require for us to study the dynamics of RNA. Lab Website Michigan Research Experts Profile |
| Andrew | Ludlow | Kinesiology | Kinesiology | Broadly, my laboratory studies telomere and telomerase biology. We are interested in the gene expression regulation of telomere maintaining genes, such as telomerase component hTERT. We focus on the gene expression regulation by alternative RNA splicing by studying deep intronic elements and the RNA binding proteins that bind these elements in diverse tissues and contexts such as in lung cancer and in cells/tissues following interventions (exercise, diet, and chemotherapy). Lab Website Michigan Research Experts Profile Featured scientist |
| Carey | Lumeng | Medical School | Pediatrics | Adipose tissue gene expression and single cell approaches. Faculty Profile Michigan Research Experts Profile Lab Website |
| Rahul | Mannan | Medical School | Pathology | Biomarker Discovery in Cancer Website Michigan Research Experts Profile |
| Anna | Mapp | LSA | Chemistry | The Mapp research group uses the tools of synthetic organic chemistry, biochemistry, and molecular biology to better understand how genes are regulated. Lab Website Google Scholar |
| David | Markovitz | Medical School | Int Med Infectious Diseases | The Markovitz laboratory studies the genetics of human centromeres, a last frontier in genomics. As part of this effort, the group investigates the RNA transcripts that arise from individual centromeres. Lab Website Michigan Research Experts Profile |
| Hayley | McLoughlin | Medical School | Neurology | Our goal is to establish pathogenic mechanisms and therapeutic interventions for neurodegenerative diseases with a particular emphasis on diseases that cause ataxia. Our work uses cell culture and transgenic mouse models as well as human tissue/biofluid samples to study neurodegenerative diseases. Our expertise includes surgical intervention, genetics, confocal imaging, and molecular and cellular biology techniques. Website |
| Miriam | Meisler | Medical School | Human Genetics | We are working on alternative splicing of sodium channel gene transcripts. Visit Lab Website Michigan Research Experts Profile |
| Daniela | Mendonca | Dentistry | Biologic & Mat Science | miRNA involved in cancer Website |
| Gustavo | Mendonca | Dentistry | Biologic & Mat Science | Effect of nano-topography on mesenchymal stem cells and differentiation into osteoblasts and the molecular basis of dental implants osseointegration. Use of RNA-Seq for identification of expressed mRNAs and miRNAs during bone healing. Website |
| Rajasree | Menon | Medical School | Comp Med and Bioinformatics | Alternative splice isoforms; Single cell RNA Website Michigan Research Experts Profile |
| Ryan | Mills | Medical School | Comp Med and Bioinformatics | Our laboratory develops methods for analyzing RNA sequence data, with a focus on quantifying and assessing rates of translation using ribosome footprinting techniques. We are also actively studying novel mechanisms of translational regulation at the transcriptional level. Lab Website Michigan Research Experts Profile Featured scientist |
| John | Moldovan | Medical School | Human Genetics | I study human LINE-1 elements, which are the only active self-mobilizing transposons in the human genome. LINE-1 proteins interact with their encoding LINE-1 RNA to form LINE-1 ribonucleoprotein (RNP) complexes. I would like to know how LINE-1 RNPs function and what host cell proteins are involved in LINE-1 mobility. Website Michigan Research Experts Profile Featured scientist |
| Stephanie | Moon | Medical School | Human Genetics | Our group is interested in how genes are expressed via the coordinated regulation of messenger RNAs at the levels of translation, localization, and decay. We study mRNA regulation in the context of human disease and stress, with a particular interest in neurological disorders. Our research aims to reveal the underlying principles and mechanisms governing mRNA in both health and disease to elucidate new therapeutic and diagnostic strategies. Lab Website Michigan Research Experts Profile Featured scientist and video |
| John | Moran | Medical School | Human Genetics | The goal of our laboratory is to understand how an abundant class of “jumping genes,” known as LINE-1 retrotransposons, impacts the structure and function of human genomes. We use molecular biological, biochemical, modern genomic, and computational approaches to address the following questions: 1) What is the molecular mechanism of LINE-1 retrotransposition? 2) How do LINE-1 retrotransposition events impact the human genome? 3) Which cellular factors promote or restrict LINE-1 retrotransposition? 4) How does LINE-1 retrotransposition contribute to intra- and inter-individual genetic variation? Website Michigan Research Experts Profile |
| Benjamin | Murdock | Medical School | Neurology | My primary work examines the role of the immune system in driving disease progression in amyotrophic lateral sclerosis (ALS). Website Lab Website |
| Deepak | Nagrath | Engineering | Biomedical Engineering | We are very interested in understanding functional effects of exosomal cargo in tumor microenvironment. In our recent work published in eLife, we uncovered the metabolic role of exosomes secreted by tumor microenvironment in cancer metabolism. Lab Website Michigan Research Experts Profile |
| Sunitha | Nagrath | Engineering | Chemical Engineering | We are developing microfluidic devices for isolating and studying circulating tumor cells (CTCs) as related to metastasis, the cause of over 90% of cancer related deaths. Lab Website Michigan Research Experts Profile |
| Jayakrishnan (JK) | Nandakumar | LSA | Molecular, Cellular, and Developmental Biology | We are interested in the biogenesis, trafficking, recruitment to telomeres, and catalytic mechanism of the RNP enzyme telomerase. We are also investigating other noncoding RNAs that are involved in gene regulation. Lab Website Michigan Research Experts Profile Featured scientist |
| Nouri | Neamati | Pharmacy | Medicinal Chemistry | The major focus of our research is to combine chemoinformatics and bioinformatics technologies to better elucidate the mechanisms of action of novel anticancer drugs developed in our laboratory. Website Michigan Research Experts Profile Lab Website |
| Alexey | Nesvizhskii | Medical School | Computation Medicine & Bioinformatics | Our research interests are in the fields of proteomics and integrative bioinformatics. On the bioinformatics side, we are focusing on the development of computational methods and tools for processing and extracting biological information from complex biological datasets. This includes computational and statistical methods for mass spectrometry-based proteomics, interactome analysis using affinity purification mass spectrometry (AP-MS) technology, proteogenomics, metabolomics, global analysis of RNA-Seq transcriptome and proteome profiles, and multi-omics data integration (RNA-Seq, proteomics, genomics, etc.) for reconstruction of pathways deregulated in cancer. Our lab has developed and maintains many computational resources widely used by the proteomics community worldwide. Website Bio Lab Website Google Scholar |
| Rachel | Niederer | Medical School | Biological Chemistry | Translation control of gene expressions plays an essential role during development, response to stress and a wide range of cellular processes. However, the key mRNA features that distinguish efficiently translated from poorly translated mRNAs remain largely unknown. Our lab utilizes a combination of high throughout experimental methods, biochemistry and molecular biology to both discover translational control elements and characterize novel regulatory mechanisms impacting gen expression. Website Michigan Research Experts Profile Lab Website |
| Erik | Nielsen | LSA | Molecular, Cellular, and Developmental Biology | We study how membrane compartments and trafficking pathways reorient during differentiation in plants. Our main model system is the differentiation of root hair cells in Arabidopsis thaliana. Lab Website |
| Roomi | Nusrat | Medical School | Internal Medicine | Use of Epic EMR for advancing care, bone marrow transplant patients, and detection of lung injury in ambulatory patients Website |
| Melanie | Ohi | Medical School | Cell & Dev Biology | The Ohi lab’s research interest focuses on dissecting the structural organization of large macromolecular machines. Our goal is to use structure as a springboard to understand function within the context of cells. Lab Website Michigan Research Experts Profile |
| Gilbert | Omenn | Medical School | Human Genetics | My RNA-related research is focused on differential expression of splice transcripts and proteins in proteogenomic studies of cancers, including a special emphasis on ERBB2+ breast cancers. Splicing is a remarkable phenomenon of the evolution of multicellular organisms with multi-exonic genes, making standard reports of up-regulation and down-regulation of “genes” a crude summary of a mixture of functionally-differentiated products of individual genes. Specific isoforms may be better biomarker candidates and therapeutic targets. Website Michigan Research Experts Profile |
| Akira | Ono | Medical School | Microbiology and Immunology | We are interested in the roles and/or behaviors of viral RNAs and cellular RNAs during virus particle assembly of HIV-1 and influenza A virus. Specific topics we are currently studying are 1) tRNA-mediated regulation of HIV-1 Gag membrane binding, 2) effects of RNA-mediated Gag multimerization on Gag localization, and 3) delivery of viral RNA segments to the site of influenza virus assembly. Lab Website Michigan Research Experts Profile |
| Edgar | Otto | Medical School | Int Med Nephrology | We apply high-throughput droplet-based single cell RNA-Seq microfluidic technology (Drop-Seq) to analyze kidney cells derived from healthy or diseased kidney samples. This project may help to identify new cell type specific biomarkers for diagnostics, disease progression, or for potential therapeutic interference. Website Michigan Research Experts Profile |
| Bruce Allan | Palfey | Medical School | Biological Chemistry | Enzyme mechanisms and kinetics. Website Michigan Research Experts Profile |
| Stephen | Parker | Medical School | Comp Med and Bioinformatics | Integration of genome, epigenome, and transcriptome (mRNA, eRNA) data across tissues and species to identify mechanisms underlying T2D GWAS SNPs. Lab Website Michigan Research Experts Profile Featured scientist |
| Abhijit | Parolia | Medical School | Pathology | Studies the role of chromatin in cancer and how its architecture may be exploited to develop new therapies. Chromatin is the tightly wound package of DNA and proteins in the nucleus of every human cell that drives transcription and gene expression. Website |
| Henry | Paulson | Medical School | Neurology | Our lab explores the reasons why the aging brain degenerates in neurodegenerative diseases, including Alzheimer’s Disease, Frontotemporal Dementia and polyglutamine expansion disorders such as Huntington disease and Spinocerebellar Ataxia type 3 (SCA3). We pursue basic studies of disease mechanisms and translational studies that are leading toward therapies for these fatal diseases. Lab Website Michigan Experts Profile |
| Bambarendage (Pini) | Perera | Public Health | Environmental Health Sciences | My primary research interests lie in the field of epigenetics, specifically in the role of environmental exposures at critical time points in life, which influence late-onset disease outcomes via epigenetic mechanisms, including aberrant DNA methylation, genomic imprinting, and small non-coding RNA (piRNA). Website Lab Website Michigan Experts Profile |
| Alexandra | Piotrowski-Daspit | Michigan Medicine | Biomedical Engineering & Internal Medicine | My laboratory develops polymeric biomaterials for therapeutic nucleic acid delivery, including a variety of RNA biomolecules. Please find more information on our website. Lab Website Michigan Research Experts Profile Michigan Faculty Profile |
| Sethu | Pitchiaya | Medical School | Urology | Interested in investigating the potential of combining anti-lncRNA therapy with existing treatment options to identify effective combinatorial treatment strategies and to further understand the global impact of ARlnc1 in prostate cancer. Faculty Profile Michigan Research Experts Profile Featured scientist Lab Website |
| John | Prensner | Medical School | Pediatrics | The Prensner lab is interested in cancer genomics, gene regulation, gene discovery, and RNA translation. Using cancer models, we study the translation of non-canonical open reading frames as both a mechanism of gene regulation as well as protein-coding functional units. We are interested in how cancer oncogenes alter RNA translational fidelity and regulation. We evaluate potential therapeutic approaches for the treatment of cancer based on aberrant RNA translation. We work within the Chad Carr Pediatric Brain Tumor Center and have a particular focus on childhood brain cancers. Lab Website View Michigan Research Experts Profile Faculty Profile |
| Jay Brito | Querido | Medical School | Biological Chemistry | The role of RNA helicases in the regulation of mRNA translation. Lab Website Michigan Research Experts Profile |
| Indika | Rajapakse | Medical School | Comp Med and Bioinformatics | The work in our lab is at the interface between mathematics and human genome biology. In particular, we study the dynamics of genome organization in human cells. From these data, our goal is to understand genome reprogrammability (=controllability). Lab Website Michigan Research Experts Profile |
| Rajesh | Rao | Medical School | Ophthalmology & Visual Science | We undertake studies related the m6a RNA methylation in retinal diseases, such as diabetic retinopathy, as well as those related to stem cell biology and retinal development. We are focused on cross-talk of the epigenome with epitranscriptomic regulators. Lab Website Michigan Research Experts Profile Faculty Profile |
| Diane | Robins* *Member Emerita | Medical School | Human Genetics | We study the role of androgen receptor in prostate cancer, using approaches such as RNA-seq in mouse and cell models to profile differential gene expression and alternative regulatory networks in disease progression and response to therapy. In a second project, we study the role of KRAB zinc finger proteins in epigenetic control of sex-specific gene expression, particularly as it impacts metabolism, in order to gain insight on the evolution of the recently expanded huge KRAB-ZFP gene family. Website Michigan Research Experts Profile |
| Anthony | Rosenzweig | Medical School | Internal Medicine | We are interested in noncoding RNAs as drivers and potential therapeutic targets in heart failure, and their role in mediating the protective effects of exercise. Website Michigan Research Experts Profile Michigan Experts Profile Lab Website |
| Liangyou | Rui | Medical School | Molecular and Integrative Physiology | RNA modifications and RNA-binding proteins, including m6A writers and readers. Obesity, diabetes, and liver disease Lab Website Faculty Profile Google Scholar |
| Brandon | Ruotolo | LSA | Chemistry | Our research is focused on developing new tools and methods aimed at determining the three dimensional structure and stability of proteins and multi-protein assemblies important in biology and medicine. Most of our technology development centers on ion mobility-mass spectrometry (IM-MS), which is an approach that can measure both the mass and the size of biomolecules in the gas phase. Our long term goal is to develop new and innovative approaches that integrate IM-MS with other structural biology tools and solve some truly challenging problems in the field of structural genomics Lab Website Faculty Profile Google Scholar |
| Russell | Ryan | Medical School | Pathology | I study distal enhancer regulation in cancer. I’m interested in learning more about the role of noncoding RNAs in enhancer function. Lab Website Michigan Research Experts Profile |
| Maureen | Sartor | Medical School | Comp Med and Bioinformatics | My lab has developed methods for the interpretation of high-throughput gene expression (e.g. RNA-seq) data in terms of pathways and processes, and we are experts in interpreting the results of such experiments with complex designs. We also study RNA in relation to HPV-related head and neck squamous cell carcinomas, and HPV-host fusion transcripts. Lab Website Michigan Research Experts Profile |
| Laura | Scott | Public Health | Biostatistics | Analysis of RNA-Seq on 300 muscle and adipose tissue samples from Finnish individuals. Website Michigan Research Experts Profile Featured scientist |
| Jiaqi | Shi | Medical School | Pathology | Our lab is interested in studying how epigenetic alterations in pancreatic cancer affect transcription and how this effect contributes to pancreatic cancer development. Website Michigan Research Experts Profile Featured scientist |
| Lyle | Simmons | LSA | Molecular, Cellular, and Developmental Biology | We study the formation and resolution of RNA:DNA hybrids. We are interested in how the cell removes ribonucleotide misincorporation events and in understanding how persistent R-loops affect genome integrity. Lab Website |
| Geoffrey | Siwo | Medical School | Internal Medicine | Understanding the origins of genetically coded proteins using AI/ Natural Language Processing CRISPR/Cas systems engineering Small molecule modulation of RNA/ nucleic acid sensing pathways. Website Michigan Research Experts Profile Internal Medicine Website |
| Janet | Smith | Medical School | Biological Chemistry | We use structural biology to study RNA viruses, including the positive-sense RNA flaviviruses (dengue, Zika, West Nile) and the HIV-1 retrovirus. We have also investigated RNA-protein interactions in Rift Valley fever virus, a negative-sense bunyavirus. Lab Website Michigan Research Experts Profile |
| Cristiane | Squarize | Dentistry | Periodontics & Oral Med | In my laboratory of Epithelial Biology, we investigate the function of key molecular signaling pathways and the contribution of stem cells to epithelial regeneration and diseases present in the skin and oral mucosa. My research focuses on answering critical clinical questions and problems related to the healing of oral mucosa and skin, in addition to studying epithelial malignancies, particularly head and neck cancer. Lab Website Michigan Research Experts Profile |
| Jeanne | Stuckey | Medical School | Biological Chemistry | Jeanne Stuckey is a member of the crystallography core of the Center for HIV RNA Studies (CRNA). She is involved in the structure determination of RNA-protein complexes important in HIV replication. Lab Website View Michigan Research Experts Profile |
| Michael Mark Alexander | Sutton | Medical School | Molecular & Behav Neurosci Inst | The remarkable information processing capacity of neurons in the mammalian brain stems from the intricate organization of their synaptic connections and the ability of these synapses to change with experience. In brain areas such as the hippocampus, the ability of principal neurons to accommodate dense networks of synaptic connectivity is critical for higher cognitive functions such as learning and memory. Our group is interested in the molecular mechanisms that control the development, maintenance and plasticity of these synapses, and in particular, how the compartmentalization of these processes in dendrites can service the unique demands of different synaptic sites impinging on the same neuron. Our research uses a combination of electrophysiology, biochemistry, and molecular approaches in conjunction with high-resolution imaging in living neurons to study these questions. Website Michigan Research Experts Profile |
| Andrew | Tai | Medical School | Int Med Nephrology | The major interest of our laboratory is to identify and characterize cellular factors that support infection by RNA viruses, chiefly hepatitis C and dengue viruses. Defining these host factors may lead to targets for broadly acting antiviral agents with high genetic barriers for resistance. Lab Website Michigan Research Experts Profile |
| Alice | Telesnitsky | Medical School | Microbiology and Immunology | The Telesnitsky lab is a virology lab that studies the genomic RNA of retroviruses. We collaborate with biophysicists to understand how alternate RNA structures define fates, and perform molecular genetics of retroviral RNA trafficking and replication roles. Lab Website Michigan Research Experts Profile |
| Muneesh | Tewari | Medical School | Int Med Nephrology | The Tewari lab studies extracellular RNA, including microRNA, long noncoding RNA and other RNA classes that are released by cells upon cell turnover or via active release (eg extracellular vesicles). We are interested in new technologies for studying extracellular RNA (eg advanced next gen sequencing and computational approaches) and are investigating extracellular RNAs in blood as non-invasive biomarkers for cancer and other diseases. Website Michigan Research Experts Profile |
| Peter | Todd | Medical School | Neurology | Our lab studies how nucleotide repeat expansions lead to neurological disease with an aim of developing novel treatments based on mechanistic insights. Specifically, we study how repeats as RNA bind to proteins and prevent them from performing their normal functions. We also study how repeats support translation of toxic proteins in the absence of an AUG start codon through a process known as RAN translation. Lab Website Michigan Research Experts Profile Featured scientist |
| Peter | Toogood | Pharmacy | Medicinal Chemistry | The goal of our medicinal research is to discover new biologically active molecules for the potential treatment of challenging diseases such as pancreatic cancer and myalgic encephalomyelitis/chronic fatigue syndrome. Our approach employs computational chemistry methods, as well as batch and continuous flow organic synthesis. Lab Website Michigan Research Experts Profile |
| Raymond | Trievel | Medical School | Biological Chemistry | Our laboratory studies the structures and functions of enzymes that regulate mRNA stability to govern gene expression and cellular metabolism. Website Michigan Research Experts Profile Lab Website |
| Natalie | Tronson | LSA | Psychology | My research focuses on the molecular mechanisms – including transcription and translation – underlying memory storage, and the modulation of memory by stress and illness. Lab Website Faculty Profile Google Scholar |
| Lam C. (Alex) | Tsoi | Medical School | Dermatology | Dr. Tsoi has strong interests in investigating the pathology and genetic architecture of complex cutaneous disorders using systems biology approaches. Working with Drs. James T. Elder and Johann E Gudjonsson, Dr. Tsoi’s research aims to develop analysis pipelines and computational approaches to provide biological inferences from genetics and genomics data. His work in genetic association studies revealed over 30 novel psoriasis susceptibility regions, and highlighted different disease pathways. He also led the analysis and developed computational pipeline to study psoriasis transcriptomes, and his work uncovered over 1,000 novel transcripts in skin. Website Michigan Research Experts Profile |
| David | Turner | Medical School | Molecular & Behav Neurosci Inst | The Turner lab studies microRNAs, gene regulation, and cell identity during development of the mammalian nervous system. We use the mouse retina as a model system and apply sequencing methods (small RNA-seq, PAR-CLIP, RNA-seq, m6A-seq), as well as CRISPR/Cas9, RNAi, and other functional methods to study or modify gene expression in differentiating neurons. Visit Website Michigan Research Experts Profile |
| Sarah | Veatch | LSA | Biophysics | We aim to understand how cells exploit the unique mixing of lipids and proteins within the plasma membrane to interpret environmental cues. Our experimental approach is rooted in a thermodynamic understanding of liquid-liquid phase separation within complex membranes. We apply and develop quantitative measures of membrane heterogeneity and protein function in living cells and model systems. Lab Website Faculty Profile Google Scholar |
| John | Voorhees | Medical School | Dermatology | Dr. Voorhees’ research focuses on psoriasis and premature aging of the skin. The demonstration of psoriasis as a disease mediated by an overactive immune system, treatable by immunosuppressive drugs, has been his major accomplishment in the study of psoriasis. In sun-induced premature skin-aging and in natural aging, he and his colleagues have unraveled mechanisms whereby UV light and the passage of time destroy the skin’s collagen support. This understanding has provided insight into its treatment and prevention by pharmacologic agents. Website Michigan Research Experts Profile |
| Nils | Walter | LSA | Chemistry | The Walter lab studies both non-coding and protein-coding RNAs, and how the former control the gene expression of the latter, using tools from biophysics, biochemistry, cell biology, molecular biology and chemical biology. Most prominently, we use leading-edge single molecule and super-resolution fluorescence microscopy and single molecule FRET approaches to probe the diverse functional mechanisms of transcriptional and translational riboswitches, the spliceosome, the RNA silencing and RNA interference machinery, ribozymes, as well as devices from DNA nanotechnology, in vitro and in live cells. Lab Website Michigan Research Experts Profile Google Scholar |
| Stanley | Watson | Medical School | Psychiatry | The emotional regulation of complex behaviors such as reinforcement, feeding and stress are central to the proper functioning of both animals and humans. The Watson laboratory focuses on the central nervous system (CNS) circuits and cellular systems that participate and regulate these states in the brains of individuals with severe mental illness. Website Michigan Research Experts Profile |
| Chase | Weidmann | Medical School | Biological Chemistry | We leverage sequencing technologies to characterize RNA-protein interaction networks in biology and human disease, with a focus on long noncoding RNAs that drive cancer metastasis. Our vision is to develop novel therapeutic strategies that target the RNA-protein interface. Website Lab Website Video |
| Xiaoquan (William) | Wen | Public Health | Biostatistics | My current research interests include topics in Bayesian model comparison, Bayesian multiple hypothesis testing and probabilistic graphical models. In applied field, I am particularly interested in seeking statistically sound and computationally efficient solutions to scientific problems in areas of genetics and functional genomics. Website Google Scholar |
| Max | Wicha | Medical School | Int Med Nephrology | The Wicha lab has been a leader in the field of cancer stem cells(CSC’s). They provides the initial description of CSC in a solid tumor are elucidating pathways which regulate these cells. This includes the study of Ln RNA’s as well as transcriptional regulators. They are also investigating epigenetic regulation of CSC and the role of chromatin structure in the regulation of the CSC transcriptome. Lab Website Website |
| Andrzej | Wierzbicki | LSA | Molecular, Cellular, and Developmental Biology | We study how non-coding RNA regulates genome activity in eukaryotes using Arabidopsis thaliana as a model system. We are particularly interested in mechanisms used by long non-coding RNA to establish repressive chromatin modifications. Website Lab Website |
| Krista | Wigginton | Engineering | Civil & Environmental Engineering | Our lab works on developing applications of biotechnology in drinking water and wastewater treatment with a focus on detecting and analyzing emerging pathogens (including RNA viruses) and their nucleic acids. We specialize in RNA detection and quantification, RNA photolysis and oxidation, and RNA environmental fate and reactivity. Lab Website Michigan Research Experts Profile |
| Ryan | Wilcox | Medical School | Internal Medicine | We have shown that GATA-3 is a proto-oncogene in T-cell lymphomas. GATA-3, among other things, regulates rRNA expression. Other novel targets currently under evaluation in the lab (e.g. CDK9) likely regulate key target genes in an RNA-dependent manner (e.g. R-loop formation at rDNA intergenic sites). Website Michigan Research Experts Profile |
| Thomas | Wilson | Medical School | Pathology | RNA-related research in the Wilson lab centers on two main areas: (1) the mechanisms by which replication-transcription conflicts impact DNA repair and genomic instability, especially copy number variants, and (2) participation with Dr. Ljungman in application of nascent RNA sequencing (Bru-seq). Lab Website Michigan Research Experts Profile |
| Patricia (Trisha) | Wittkopp | LSA | Ecology and Evolutionary Biology; Molecular, Cellular, and Developmental Biology | We study the genetic mechanisms controlling RNA production and how they vary within and between species. Lab Website Michigan Research Experts Profile Featured scientist |
| Connie | Wu | Engineering | Biomedical Engineering | Our lab works at the interface of biology, bioanalytical chemistry, and materials and biomolecular engineering to develop technologies that can address diagnostic and therapeutic challenges related to human health. Our long-term translational goals are to accelerate biomarker signature discovery for applications in cancer and to engineer multifunctional RNA therapeutics and rationally designed nanomaterials. Lab Website Website Google Scholar |
| Swathi | Yadlapalli | Medical School | Cell and Developmental Biology | We are focused on understanding the molecular and cellular mechanisms controlling circadian rhythms and sleep. We are particularly interested in molecular mechanisms governing post-transcriptional splicing and mRNA localization to cellular organelles. Lab Website Website Google Scholar |
| Bing | Ye | Medical School | Cell & Dev Biology | mRNA processing, translation, and RNA-binding proteins in neuronal development. We study post-transcriptional controls and RNA-binding proteins in the development of dendrites and axons Lab Website Website Google Scholar |
| Chengxin | Zhang | Medical School | Computational Medicine & Bioinformatics | RNA structure and function prediction. The principal goal of our research is to reveal the fundamental relationship between protein sequence, structure and function Website Michigan Experts Profile Google Scholar |
| Jianzhi (George) | Zhang | LSA | Ecology and Evolutionary Biology | RNA modification, RNA folding, translation Lab Website Featured scientist |
| Jifeng | Zhang | Medical School | UMH Internal Med-Cardiovascular | Dr. Jifeng Zhang is Professor of cardiovascular medicine at the University of Michigan. He is among the first group of researchers to develop and use the ZFN, TALEN, and CRISPR/Cas9 technologies to generate gene knock-out and knock-in animal models, including mice, rabbits, and pigs, for translational research. His team has contributed over 30 rabbit models to date, including MPO, TRIM5, ADCY9, CETP, ApoBEC1, IDOL, apoAII, apoE, and ApoCIII KO, to name just a few, made available to the community for translational medicine and drug development in various fields of human disease study. Michigan Research Experts Profile Website |
| Yan | Zhang | Medical School | Biological Chemistry | Research interests include CRISPR, Cas9, NmeCas9, CRISPR-Cas3 system. Lab Website Michigan Research Experts Google Scholar Website |
| Yue | Zhao | Medical School | Comp Med & Bioinformatics | My research focuses on developing and applying statistical methods for high-dimensional data integration in biomedical research. I have collaborated with basic science researchers and physicians to design optimal multi-omics data collection and perform advanced analyses, such as genomics, transcriptomics, proteomics, metabolomics, CRISPR-based genetic screening, and third-generation sequencing. My research goals include investigating the effects of environmental exposures on genome, epigenome, transcriptome, and epi-transcriptome in neurodegenerative diseases like ALS, as well as conducting multi-omics analysis to establish connections between exposures, omics, and disease risk. Website Google Scholar |
| Xiang | Zhou | Public Health | Biostatistics | We are a group of statisticians and computational biologists developing statistical methods and computational tools for genetic and genomic studies. These studies often involve large-scale and high-dimensional data; examples include genome-wide association studies and various functional genomic sequencing studies such as bulk and single cell RNA sequencing, bisulfite sequencing, and spatially resolved transcriptomics. By developing novel analytic methods, we seek to extract important information from these data and to advance our understanding of the genetic basis of phenotypic variation for various human diseases and disease related quantitative traits. Lab Website Michigan Research Experts Profile Website Google Scholar |
| Guizhi (Julian) | Zhu | Medical School | Pharmaceutical Sciences | With the overarching goal to develop novel and clinically translatable nucleic acid immunotherapeutics and vaccines, Zhu Lab’s research interest lies at the interface of RNA/DNA/protein chemistry and engineering, immunology, and applied chemistry and biomaterials. Our current focus is to develop circRNA/mRNA/oligonucleotide immunotherapeutics/vaccines and their delivery systems for the prophylaxis or immunotherapy of cancer, infectious diseases, and autoimmune disorders. Lab Website Michigan Research Experts Profile College of Pharmacy Faculty Profile |
