What is RNA?
RNA, or ribonucleic acid, represents a major determinant of cellular life. The promise of the ~$3-billion human genome project around the turn of the millennium was to pave the way for the genetically personalized medical treatment of so far intractable human diseases such as cancer and Alzheimer’s. Instead, it revealed that the overwhelming majority – perhaps >90% – of our genome encodes often highly conserved RNA molecules, compared with <2% that encodes – via messenger RNA mediators – proteins, the components previously believed to dominate cellular function. In fact, it is now understood that the human genome harbors at least 80,000 non-redundant non-coding RNA genes, a revolutionary insight that has led researchers to dub the eukaryotic cell an “RNA machine”. This profound realization – after almost a century of focusing on proteins – calls for a swift shift towards studying the plethora of emerging RNA functions in human life and disease.
What are the roles of RNA in cells?
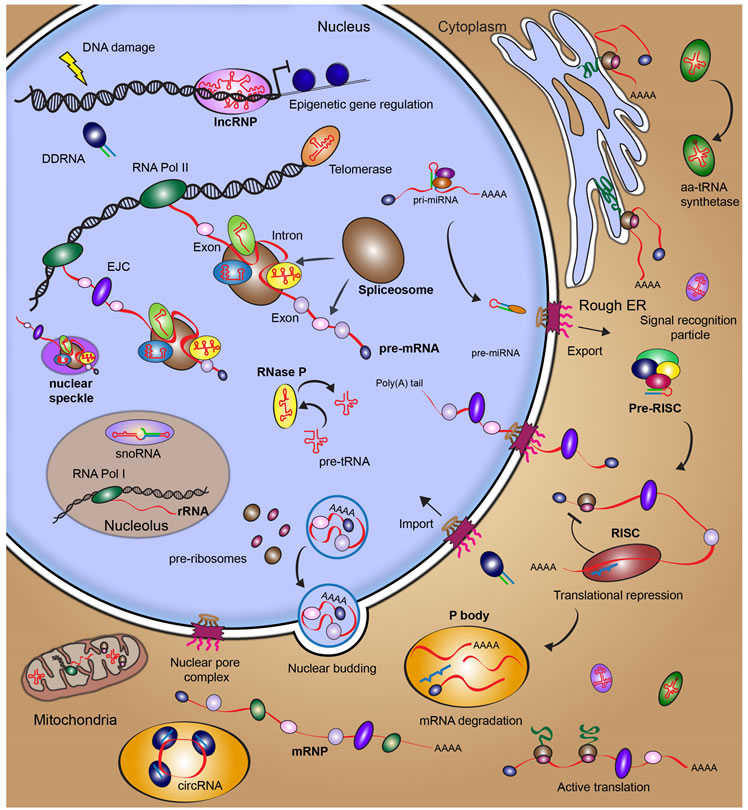
The plethora of RNAs recently identified in human cells expose an unexpectedly complex, highly interconnected RNA metabolism in our body (with excerpts of types of RNAs and associated known functions highlighted in Figure 1). This demonstrates that for a long time we have underestimated the intricacy involved in the human genome maintenance, processing and regulation performed by these RNAs. With the still growing catalogue of RNAs in hand, the time has come to strive to understand the multitude of their functions inside the human cell and fulfill the unresolved legacy of the human genome project.
How is the expression of RNA regulated?
One broad role of RNA lies in determining when how much of which protein and RNA is produced for a cell to properly function. It has long been known that a human cell harbors about 100,000 different types of proteins. It was therefore expected that the human genome project would unveil this as the number of protein-coding genes. However, only about ~20,000 coding genes were found. How could that be? It turns out that each protein-coding gene can be read in different ways and the resulting RNA “spliced” into different “isoforms” that are slight variations of the primary gene sequence. Instead of one gene encoding one protein, the average gene is read into 7 different versions, each coding for a slightly different protein with a distinct function. The introns thus spliced out from protein-coding genes provide perhaps the largest source of non-coding RNA in the cell. The levels of each RNA in each of the 100 trillion cells of the human body are under strict regulation. Each gene has a so-called “promoter element” to which specific factors bind to direct the transcription machinery to the gene to be expressed. Nearby enhancer elements, when activated, amplify this signal by generating “enhancer RNAs” in ways that are still poorly understood. Many of the disease-related sequence variations identified in our genome are found in enhancer or promoter regions, underscoring the importance of properly regulating the transcriptional output for normal physiology as well as human diversity. Furthermore, our DNA is organized into a structure called chromatin that packages about 2 meters of DNA into a sphere of about 10 micrometers diameter and controls access to segments of this DNA. The DNA as well as the packaging proteins (histones) often are chemically modified by RNA effector complexes, which regulate the activities of the genome and establish the particular identity of the cell by “epigenetics”. Finally, RNA- and protein-based regulators tightly control the lifespans and thus levels of RNAs in all cells (Figure 1).
RNA in human disease
The advent of next-generation sequencing and other high-throughput “–omics” technologies has led to increasing recognition of RNA as a key player in the pathophysiology of cancer and other diseases. Mutations in essential splicing factor genes frequently occur in several malignancies and solid tumors, whereas aberrant RNA processing has been implicated in neurodegenerative diseases, including ALS and Alzheimer’s. Altered regulation of different RNA splice forms can also contribute to disease; for example, an isoenzyme of pyruvate kinase has been shown to be necessary for cancer metabolism and tumor growth. This is an exciting time for RNA biology and human disease because such examples are likely the tip of the iceberg, with many more discoveries of RNA-based disease mechanisms sure to come.
Early detection and biomarkers
Recent discoveries have prompted heightened interest in the use of RNA as a biomarker to monitor diseases. In 2008, cell-free microRNAs were discovered in the bloodstream. Extracellular microRNAs in easily accessible body fluids hold great promise as biomarkers, as they could be used as a “liquid biopsy” for minimally invasive diagnosis or monitoring of cancer and other diseases. RNAs are particularly appealing as biomarkers because of their dynamic nature — expression of RNA and its splice forms change with physiology, and these variations could be used for earlier detection and improved monitoring of disease over time. It has also become clear that other extracellular RNA species beyond microRNAs exist in blood and other biofluids, and these too may hold promise as biomarkers.
RNA-based therapeutics
The discovery of functional RNAs (particularly small interfering and microRNAs) has led to increased efforts to develop RNA-based therapeutics. For example, a 2013 clinical trial showed that a new drug known as Miravirsen was able to block replication of the Hepatitis C virus in infected patients by inhibiting a liver-specific microRNA. Another drug, bevasiranib, is a first-in-class synthetic small interfering RNA that impedes angiogenesis for the treatment of eye conditions such as diabetic macular edema and age-related macular degeneration. Attempts at RNA delivery to other tissues have proven challenging, but ongoing research is working to overcome this hurdle. With the increasing discovery and study of RNA-based mechanisms operative in disease, we can expect many more opportunities for RNA-based therapeutics.
References
- Pitchiaya, S., Heinicke, L. A., Custer, T. C. & Walter, N. G. (2014). Single molecule fluorescence approaches shed light on intracellular RNAs. Chem Rev 114, 3224-65. PMC-ID: 3968247.
