“Kinetic fingerprinting” of single protein molecules to find the biomarker needle in the haystack
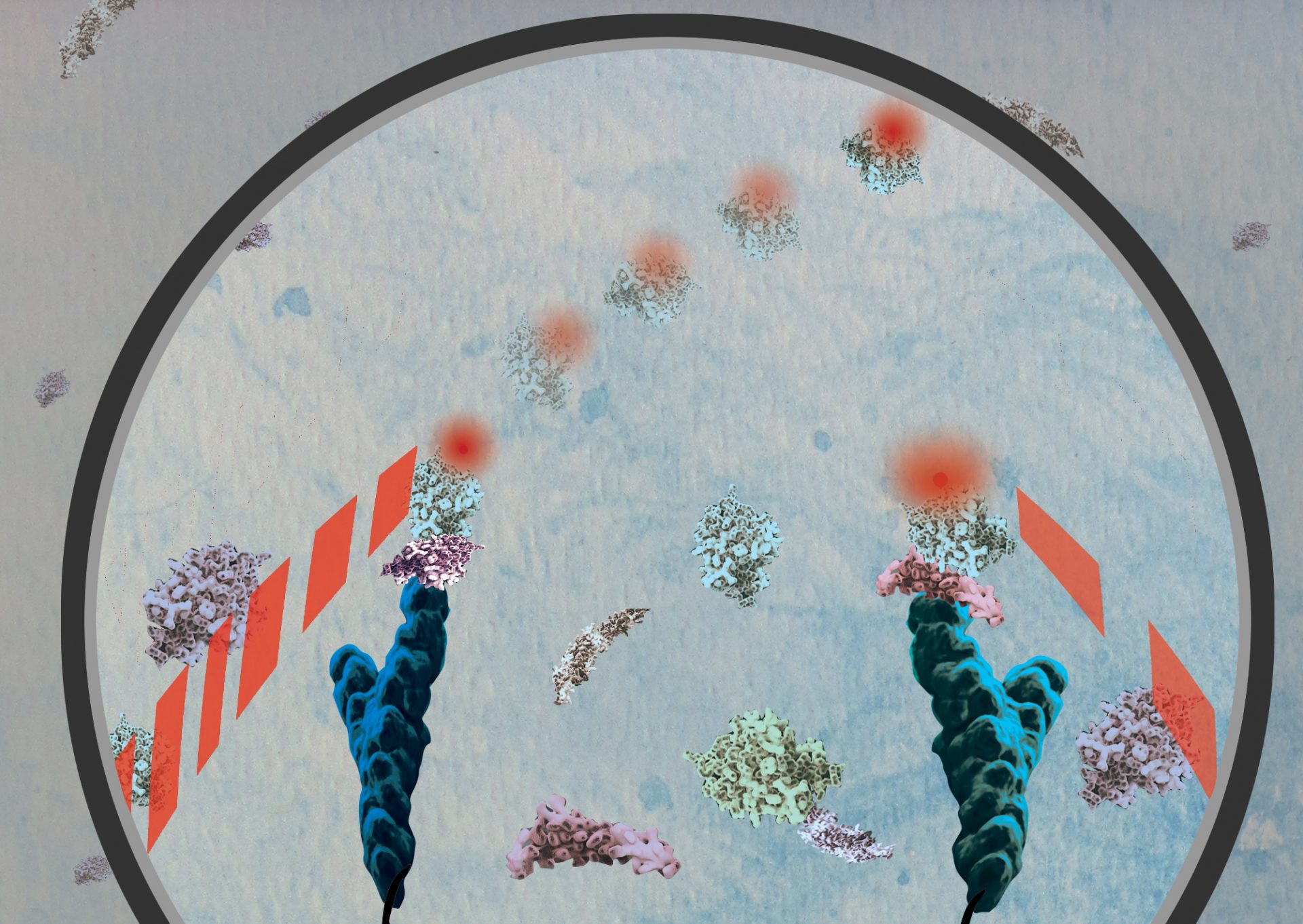
A cross-disciplinary team of scientists from the University of Michigan (U-M) has developed a biochemical technique that successfully measures the number of individual protein molecules present in blood at low concentrations. These findings are published in the Proceedings of the National Academy of Sciences (PNAS), September 2020.
The scientists developed special antibody probes that repeatedly bind to and then release the target protein. With a single-molecule fluorescence microscope, they measured the number of times and for how long the repeated binding events occurred.
The repeated binding behavior creates a specific pattern, called a “kinetic fingerprint,” that helps to positively identify the target protein. Typically, each binding event lasts for a few seconds, and the entire measurement can take a couple of minutes.
The authors reasoned that if many antibodies—say, 10 to 20—bind to the same protein molecule, there is a very high probability that the antibodies have found the target protein. If only one or two antibodies bind to/dissociate from a protein, it is likely to be a random occurrence. This realization allowed the authors to filter out the background signal that plagues conventional protein tests, resulting in more sensitive measurements.
The team initially developed kinetic fingerprinting on nucleic acids (DNA and RNA), for which probes are easier to produce than for proteins. The present work marks the first time that this technique has been used to detect proteins at much lower concentration than traditional assays.
“Finding biomarker proteins can be like finding a needle in a haystack,” explains Alexander Johnson-Buck, Ph.D., a biophysical chemist and co-author of this publication. “There can be very few target proteins in a specimen, and they can be very hard to detect.”
This technique could allow the use of many more biomarker proteins for diagnostics. “There might be many useful biomarkers of diseases that are too low in concentration to be measured with conventional assays,” explains Johnson-Buck, “and with kinetic fingerprinting, we now have the technology to find them.”
The team measured four model proteins (IL-6, IL-34, PAI-1, and VEGF-A) in blood sera, with virtually no background signal. This low background permitted detecting the proteins at concentrations approximately one hundred times lower than conventional protein assays.
In addition to high sensitivity, there are several important advantages of the kinetic fingerprinting technique over conventional assays. First, the novel technique can use antibodies that bind so weakly to their target proteins that they would be useless in conventional protein assays, which typically require tightly binding antibodies. Since weakly binding antibodies are easier to develop, this characteristic will likely reduce costs and broaden the applications for the new technique.
In addition, kinetic fingerprinting does not require washing away excess antibodies before a measurement, which is critical for accuracy in conventional assays. Avoiding these washing steps saves hands-on time and eliminates one source of variation between measurements while maintaining high sensitivity.
In the future, it is expected that with kinetic fingerprinting, scientists will be able to measure the presence of many proteins in the blood that are too dilute to be measured conventionally. This will expand the number of possible biomarker proteins for diagnostics. Since bacteria and viruses are covered with distinctive proteins, kinetic fingerprinting might also constitute a more sensitive and faster technique to test for such pathogens.
The kinetic fingerprinting technique was first developed, and early studies performed, using the Single Particle Tracker (SPT) microscope available at the U-M Center for RNA Biomedicine SMART Center. Tanmay Chatterjee, Ph.D., first author of this publication, played an essential role in collecting the data.
“The key element was getting people together with the right expertise and complementary skill sets,” says Johnson-Buck. “It was critical to have the clinical specimen collection capability of the Tewari Lab brought together with the basic science strengths of Nils Walter’s lab.”
Muneesh Tewari is the Ray and Ruth Anderson-Laurence M Sprague Memorial Research Professor, Professor of Internal Medicine, Medical School and Professor of Biomedical Engineering, Medical School and College of Engineering
Nils G. Walter is the Francis S Collins Collegiate Professor of Chemistry, Biophysics and Biological Chemistry, Professor of Chemistry, Professor of Biophysics, College of Literature, Science, and the Arts, and co-director of the U-M Center for RNA Biomedicine.
Competing financial interests
The University of Michigan has filed patent applications on technologies described herein, on which Muneesh Tewari, Nils Walter, and Alexander Johnson-Buck are listed as inventors. Drs. Tewari, Walter, and Johnson-Buck are co-founders of aLight Sciences, LLC, which sponsored this work and seeks to commercialize the SiMREPS technology.
Cited paper:
Direct kinetic fingerprinting and digital counting of single protein molecules, Tanmay Chatterjee, Achim Knappik, Erin Sandford, Muneesh Tewari, Sung Won Choi, William B. Strong, Evan P. Thrush, Kenneth J. Oh, Ning Liu, Nils G. Walter, Alexander Johnson-Buck, PNAS, 2020, https://doi.org/10.1073/pnas.2008312117

