Machine learning expands single-molecule analysis accuracy and accessibility
 The observation of single biomolecules in real-time is crucial for our understanding of the cellular biology that is assembled from these molecules, be they DNA, RNA or protein. The recent development of an array of tools and techniques for single-molecule analysis allows studies at an extremely small scale (nanometers, or 10-9 meters) over short periods of time (from a few milliseconds to a second).
The observation of single biomolecules in real-time is crucial for our understanding of the cellular biology that is assembled from these molecules, be they DNA, RNA or protein. The recent development of an array of tools and techniques for single-molecule analysis allows studies at an extremely small scale (nanometers, or 10-9 meters) over short periods of time (from a few milliseconds to a second).
However, until now, most of such observations required tedious and time-consuming manual data processing of thousands of single molecules. A team of University of Michigan (U-M) Department of Chemistry and Department of Physics scientists, spearheaded by graduate students Jieming Li, now Ph.D. in Chemistry, and Leyou Zhang, now Ph.D. in Physics, developed a deep learning algorithm to analyze data emerging from a single molecule microscope. The results from this collaboration are published in Nature Communications (November 2020).
Illustration: Deep learning assisted single molecule fluorescence microscopy data analysis
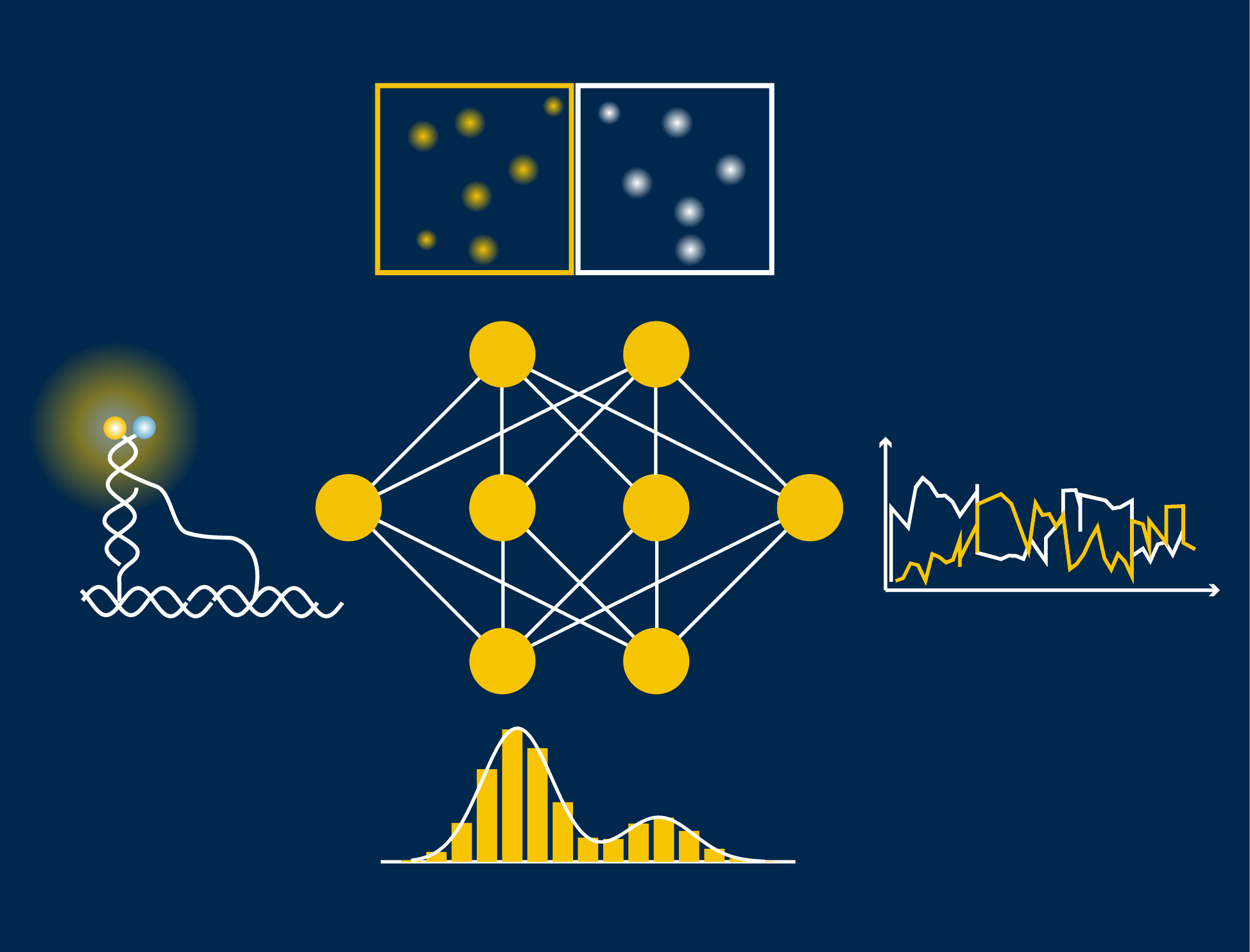
The key to their automatic data analysis workflow for single molecules (called “AutoSiM”) is to apply a deep learning algorithm, representing a special class of machine learning or artificial intelligence that contains many layers of a neural network, to single molecule fluorescence microscopy data. First, the pattern of the fluorescence signal is recognized to classify the molecule as either relevant or not. Then, the algorithm distinguishes which segment within the fluorescence signal should be included for further data interpretation. For training, the scientists fed the program with a large dataset that was already analyzed by human experts to teach the algorithm to recognize the correct pattern of fluorescence signals, which reflect the behavior of bio-molecules. Once trained on one dataset, it can quickly be adapted to any new dataset by a short “transfer learning”.
There are three main advantages to using AutoSiM: it is faster, significantly cuts down on research time, and reduces costs. “One of my goals is to free up researchers from doing tedious data analysis so they can better focus on their exciting scientific inquiries,” says Li. With the network, data analysis is more consistent than possible across human researchers (due to individual biases), and is free from data entry errors. The concordance between AutoSiM and manual selection is about 90% and is completely reproducible from day to day.
AutoSiM was developed on two datasets from the lab of Nils Walter, U-M Francis S. Collins Collegiate Professor of Chemistry, Biophysics, and Biological Chemistry. One dataset comprised kinetic fingerprints of single molecules termed SiMREPS time traces. The SiMREPS assay distinguishes surface-immobilized mutant DNA biomarkers of disease from wild-type on the basis of distinguishable fluorescence kinetics when a probe interacts with the mutant sequence, wild-type sequence, or surface itself. This molecular diagnostics tool is now being commercialized by aLight Sciences Corp., co-founded by Walter and co-author Alexander Johnson Buck. The other set consisted of time traces characterizing the shape changes of 4 different biomolecular complexes using single molecule FRET (smFRET). Since its development over two decades ago, smFRET has been used to study the dynamics of many biomolecules at the nanometer scale, especially conformational changes involving nucleic acids and/or proteins.
The workflow allows for transfer learning, which means that it can be adapted to new systems by learning from small additional datasets, expanding the original capabilities of the algorithm. “This is like our human brain that learns a lot of new information at a young age, but can also readily adopt new input over its entire lifespan,” says Walter. “It’d be great if others would include their own data to train and use AutoSiM, so the capabilities of the network can be expanded,” explains Li.
The newly released software is available and free for academic purposes, through the U-M Library Deep Blue data repository. The software comes with a basic set of instructions. The Walter lab has already received two requests to use the software. One is for multiplexing data analysis, and the other one is for nanopore data analysis.
The University of Michigan (U-M) supports state-of-the-art single molecular microscopy at the Single Molecule Analysis in Real-Time (SMART) Center, one of the two core facilities of the U-M Center for RNA Biomedicine. The SMART Center is a shared-use facility providing university researchers with single molecule detection and manipulation tools to track and analyze biomolecules with unprecedented detail. The SMART Center provides access to instrumentation, including single molecule spectroscopy and imaging, laser tweezers, and atomic force microscopy; as well as experienced support in experimental planning and analysis.
Paper cited:
Classification and segmentation of single-molecule fluorescence time traces with deep learning, Jieming Li, Leyou Zhang, Alexander Johnson-Buck and Nils G. Walter, Nature Communications, (2020)11:5833, doi.org/10.1038/s41467-020-19673-1