Researchers in the Moon Lab publish findings on how emergency response RNAs avoid condensation during stress – a discovery that could help fight disease
A new study from the University of Michigan’s Stephanie Moon Lab, with first-author Genetics and Genomics Ph.D. candidate Noah Helton, offers fresh insights into how cells manage molecular crises.
Helton, the first graduate student to join the lab, worked closely with Principal Investigator Stephanie Moon, Ph.D., Assistant Professor of Human Genetics and Center for RNA Biomedicine Faculty Scholar, and Research Scientist Benjamin Dodd. Their findings, published in Genes & Development, represent the Moon Lab’s first research paper authored exclusively by lab members — a significant milestone.
“We’ve published reviews before, but this is the first research paper from my lab written solely by Moon Lab members, and such a short author list is rare these days,” says Moon.
For several years, Moon’s research has focused on how cells respond to stress.
In healthy cells, most RNA molecules are covered with ribosomes, which function like miniature factories, translating genetic instructions in RNA into the proteins essential for cellular function.

Stephanie Moon, Ph.D., Assistant
Professor of Human Genetics, Medical School, Faculty Scholar, Center for RNA Biomedicine
When cells are stressed by heat, toxins, inflammation, or other challenges, most cellular processes, including protein production, are temporarily shut down to help the cell survive.
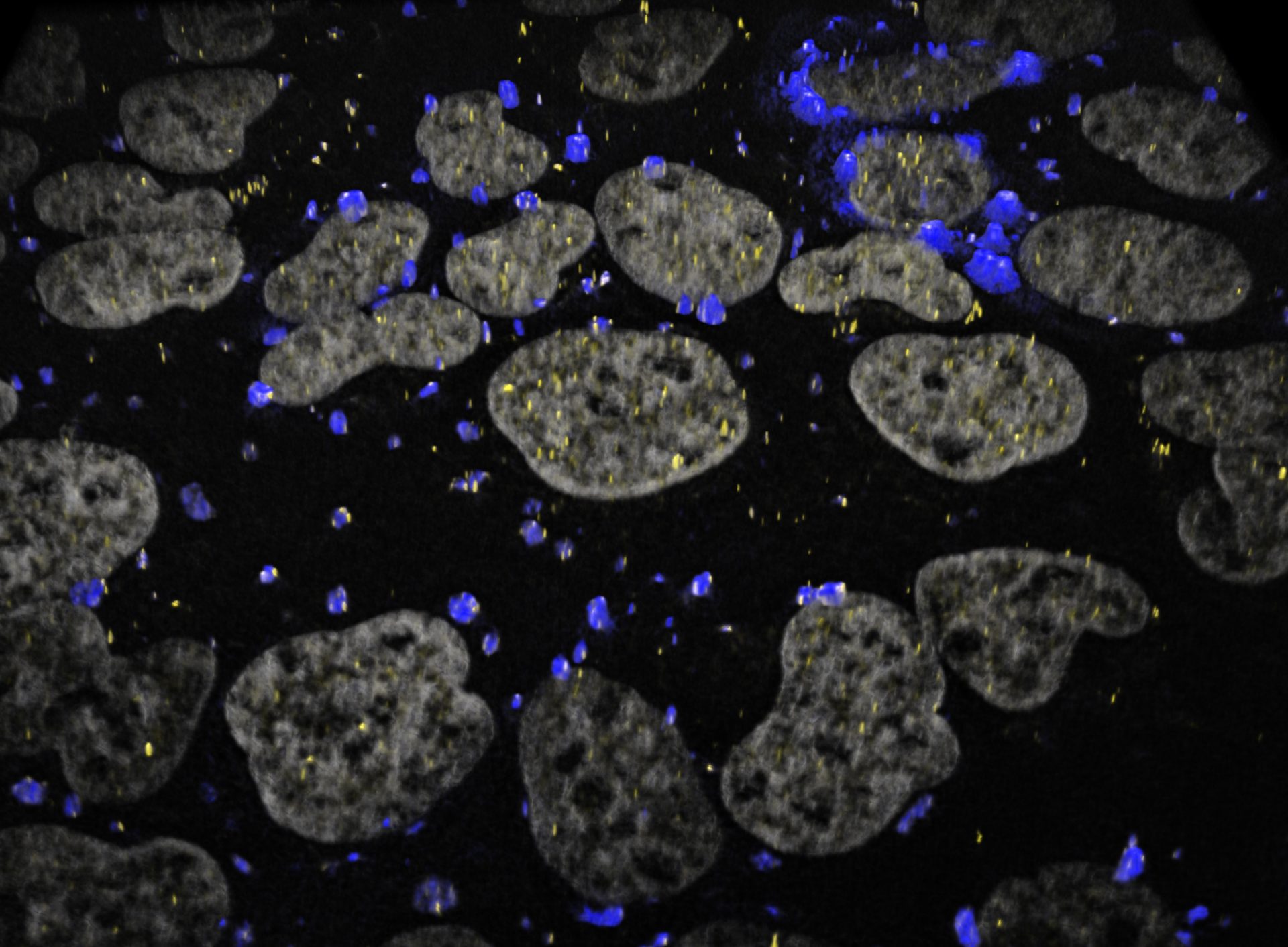
Ribosomes typically detach or “run off” RNAs as part of this response. These unprotected RNAs then clump together in blobs called “stress granules,” which act as temporary storage sites until conditions return to normal.

Genetics and Genomics Ph.D. Candidate
The Moon Lab, University of Michigan
However, certain mRNAs (messenger RNAs) need to be expressed during stress to help the cell recover and adapt.
“These specialized mRNAs are like emergency vehicles speeding toward a wreck on an interstate highway where they can quickly respond,” says Moon. “The ‘regular’ traffic (other mRNAs) is sent off the road (into granules) during the crisis.”
Before this study, it wasn’t clear if or how these specialized mRNAs were kept out of stress granules. While many studies hinted that translation of these mRNAs during stress would exclude them from stress granules, other recent studies found evidence that these mRNAs could translate inside stress granules.
The Moon Lab showed that these vital mRNAs escape stress granules by interacting with ribosomes. Trapping these mRNAs in stress granules would halt their protein production at a time when the cell may need them most.
Understanding the integrated stress response (ISR) pathway is paramount, since disruptions in stress granule dynamics are implicated in neurological diseases (including ALS), cancer, and other conditions where cells are exposed to this heightened state or “chronic” stress.
Given these stakes, the Moon Lab team posed a crucial question: What enables certain mRNAs to stay out of stress granules?
Scientists already knew that uORFs (upstream open reading frames) — “extra instructions” or special sequences found at the front, 5′ untranslated region (5′ UTR) of certain mRNAs — can promote ribosome recruitment and their attachment to mRNAs.
Helton et al., wanted to dig deeper and asked: Do uORFs help certain mRNAs keep or attract ribosomes during stress and therefore avoid the stress granule sandtrap?
To test this, Helton and the team used chemical inhibitors, single molecule imaging of cellular mRNAs, live-cell imaging with fluorescent proteins, and engineered RNA “reporters.” They confirmed that the presence of a uORF leads to greater ribosome association with mRNAs under stress.
Their experiments showed that removing the uORF resulted in a loss of ribosome association, and the RNA was then much more likely to end up in a stress granule.
Surprisingly, they also found that even just one ribosome lingering on an mRNA is enough to protect the mRNA from condensation into a granule.
Scientists had previously thought that an RNA strand needed to have multiple ribosomes attached to avoid being silenced in a stress granule. The new findings from the Moon Lab turn that theory on its head, that even a single ribosome is sufficient to keep an mRNA out of stress granules and allow it to continue making proteins.

stress granules (green) in stressed human cells (nuclei in blue).
Image courtesy of Stephanie Moon, Ph.D.
Dodd explains: “If you think of RNA as a string and the ribosome as a bead, it has been the prevailing thought that multiple beads on the string act as a ‘shield’ against the formation of stress granules because there are so many beads. However, what this paper reveals is that having just one bead — or ribosome — on the string (RNA) does the same thing.”
Furthermore, the team clarified how even a tiny, 6-nucleotide change to remove start codons from a uORF would drive an mRNA into a granule.
The fundamental insights gained from the Moon Lab’s study could ultimately help inform the development of new treatments and therapeutic interventions to maintain healthy protein synthesis in diseases where the stress response goes awry.
“We’re excited to further explore the interplay of uORFs and RNAs, and learn more about how this single ribosome can prevent mRNA from tangling and forming stress granules,” says Helton. “I’m looking forward to continuing to work with this fantastic Moon Lab team, and can’t wait to share what we find out next.”
Paper cited: “Ribosome association inhibits stress-induced gene mRNA localization to stress granules.” Genes & Development. DOI: 10.1101/gad.352899.125
Funding: Supported by National Institute of General Medical Sciences (R35GM146711) and the Chan Zuckerberg Initiative