Koutmou and Koutmos results published in the Proceedings of the National Academy of Sciences (PNAS)


A University of Michigan team of biochemists, led by Kristin Koutmou, Ph.D., and Markos Koutmos, Ph.D., Assistant Professors in the Department of Chemistry and Department of Biophysics, is reframing the understanding of the biology of a class of enzymes called Pseudouridine Synthases (Pus enzymes). These enzymes modify many types of RNAs, and the Koutmou and Koutmos Labs’ research brings new insights into the selection principles that guide modification incorporation. Their results are published in the Proceedings of the National Academy of Sciences (PNAS).
This novel understanding of pseudouridine biology could have important therapeutic implications because the dysregulation of Pus enzymes is linked to inherited diseases impacting muscle and brain function, such as progressive mitochondrial myopathy and sideroblastic anemia (MLASA). Furthermore, these enzymes also catalyze pseudouridine incorporation into RNA viral genomes, including that of SARS-CoV-2. As such, Pus enzymes present a potential new target for the development of therapeutics.
Since RNAs are central to the protein synthesis machinery, its chemical modifications can alter how fast and/or how accurately proteins are made. These modifications can have many consequences for cellular health, overall cellular adaptation, and cellular regulation.
This new PNAS publication investigates one of the most common RNA modifications, pseudouridine, which is often found in the mRNA region decoded by the ribosome. Since the 1960s, the enzymes that incorporate pseudouridine have been known to target transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs). In 2014, it was discovered that some Pseudouridine Synthases also modify mRNAs, which are structurally very different from tRNAs and rRNAs. This discovery opened the question of understanding how Pus enzymes target so many different types of RNAs.
In this publication, the Koutmou and Koutmos labs teamed together to look at Pseudouridine Synthase 7
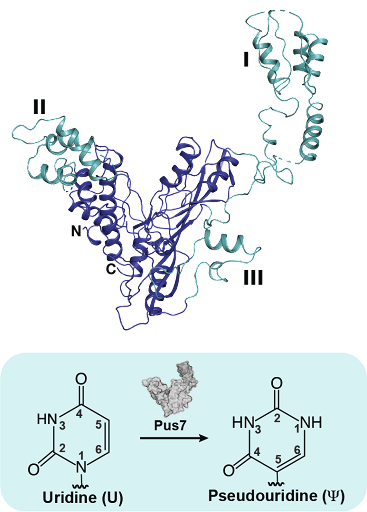
(Pus7), one of the main enzymes responsible for adding pseudouridine into mRNAs. To establish a structural basis for how RNA targets are recognized by Pus7, Markos Koutmos and Meredith Purchal, a graduate student in the Program in Chemical Biology, obtained a molecular-level picture of the yeast enzyme using X-ray crystallography. Their X-ray structure revealed that while the core of the enzyme is very similar to the bacterial homologue, Pus7 possesses extra peripheral eukaryotic insertions that might be important for the enzyme to recognize its broad set of mRNA and non-coding RNA targets.
Based on their structure and available data on similar enzymes, the team hypothesized that Pus7 selected its RNA targets by recognizing specific folded structures. “We were totally wrong,” said Koutmou. “As we performed these studies, we realized that this enzyme would modify pretty much anything that had a short consensus sequence, regardless of the RNA structure.” These results established that, to their surprise, it is not RNA structure that governs pseudouridine incorporation. This was initially confounding because Pus7 modifies its consensus sequence only 2% of the time in cells. Dr. Koutmou remarked that “As scientists, it was very frustrating because we were unable to make it not modify! These results certainly made us revisit our hypothesis.”
From the tube to the cell
What, then, makes Pus7 modify such a limited number of the possible RNAs? Unlike when purified and studied in vitro, RNAs in the cell are almost never alone. Being super negatively charged, many proteins and other molecules bind to them, potentially occluding the consensus sequence or stabilizing RNA secondary structures that render the consensus sequence inaccessible. This made the Koutmou and Koutmos teams hypothesize that Pus7 selects its targets based upon the accessibility of these sequences.
This idea launched a series of temperature-dependent and modelling studies demonstrating that Pus7 more frequently and rapidly modifies RNAs with accessible consensus sequences. These results fit with other studies showing that when exposed to heat stress, Pus7 relocalizes from the nucleus into the cytoplasm where it modifies > 200 additional RNA targets. The change in localization increases the local abundance of Pus7 enzymes in the cytoplasm, where, due to the heat shock, the enzyme is more likely to encounter mRNAs with Pus7 exposed consensus sequences. These changes are not permanent and occur in response to transient stress.
The Koutmou and Koutmos teams’ results bring a fresh perspective to understanding how some Pseudouridine Synthases might recognize their substrates by demonstrating that Pus7 substrate selections rely heavily on accessibility to the RNA targets — as opposed to a specific secondary structure. “This is really a reframing of how we think about the strategies used by this class of enzymes to select substrates,” concluded Koutmou.
The flexibility in Pseudouridine Synthase-mRNA substrate selection could be an important process for the adaptation and fitness of the cell, by which it can quickly change the protein synthesis without having to permanently change the DNA. Additional research will further explore these potential adaptive mechanisms.
Kristin Koutmou, Ph.D., is the Seyhan N. Ege Assistant Professor of Chemistry, Department of Literature, Science and the arts, at the University of Michigan.
Markos Koutmos, Ph.D., is a member of the Executive Committee for the Center for RNA Biomedicine and Assistant Professor of Chemistry and Biophysics, Department of Literature, Science and the arts, at the University of Michigan.
Written by: Elisabeth Paymal
Paper cited:
“Pseudouridine synthase 7 is an opportunistic enzyme that binds and modifies substrates with diverse sequences and structures,” Meredith K. Purchal, Daniel E. Eyler, Mehmet Tardu, Monika K. Franco, Megan M. Korn, Taslima Khan, Ryan McNassor, Rachel Giles, Katherine Lev, Hari Sharma, Jeremy Monroe, Leena Mallik, Markos Koutmos and Kristin S. Koutmou, Proceedings of the National Academy of Sciences, 1/25/2022, DOI: 10.1073/pnas.2109708119