Making patients resilient to COVID-19: Lessons to be learned from kidney single cell analysis
by Dr. Kretzler and his team, and Elisabeth Paymal, U-M Center for RNA Biomedicine
The scientific response to the COVID-19 pandemic is rapidly evolving. Every day, scientists are making discoveries based on years of investment in training and expertise that resulted in the accumulation of large, shared, high quality data sets now ready to be used in the fight against the virus. Together, researchers have established lasting scientific collaborations and built robust state of the art research infrastructures that are now strongly positioned to tackle emergent questions related to the treatment and spread of disease during this COVID-19 pandemic.

As I spoke with U-M Center for RNA Biomedicine member Matthias Kretzler, M.D., University of Michigan, Department of Nephrology/Internal Medicine and Computational Medicine and Bioinformatics, he was launching a new research protocol. “We might be able to start collecting and generating data from COVID-19 infected kidney cells as soon as tomorrow, so we can help patients with kidney diseases and those with COVID-19 induced kidney damage. The sooner we can start, the sooner we can find answers.”
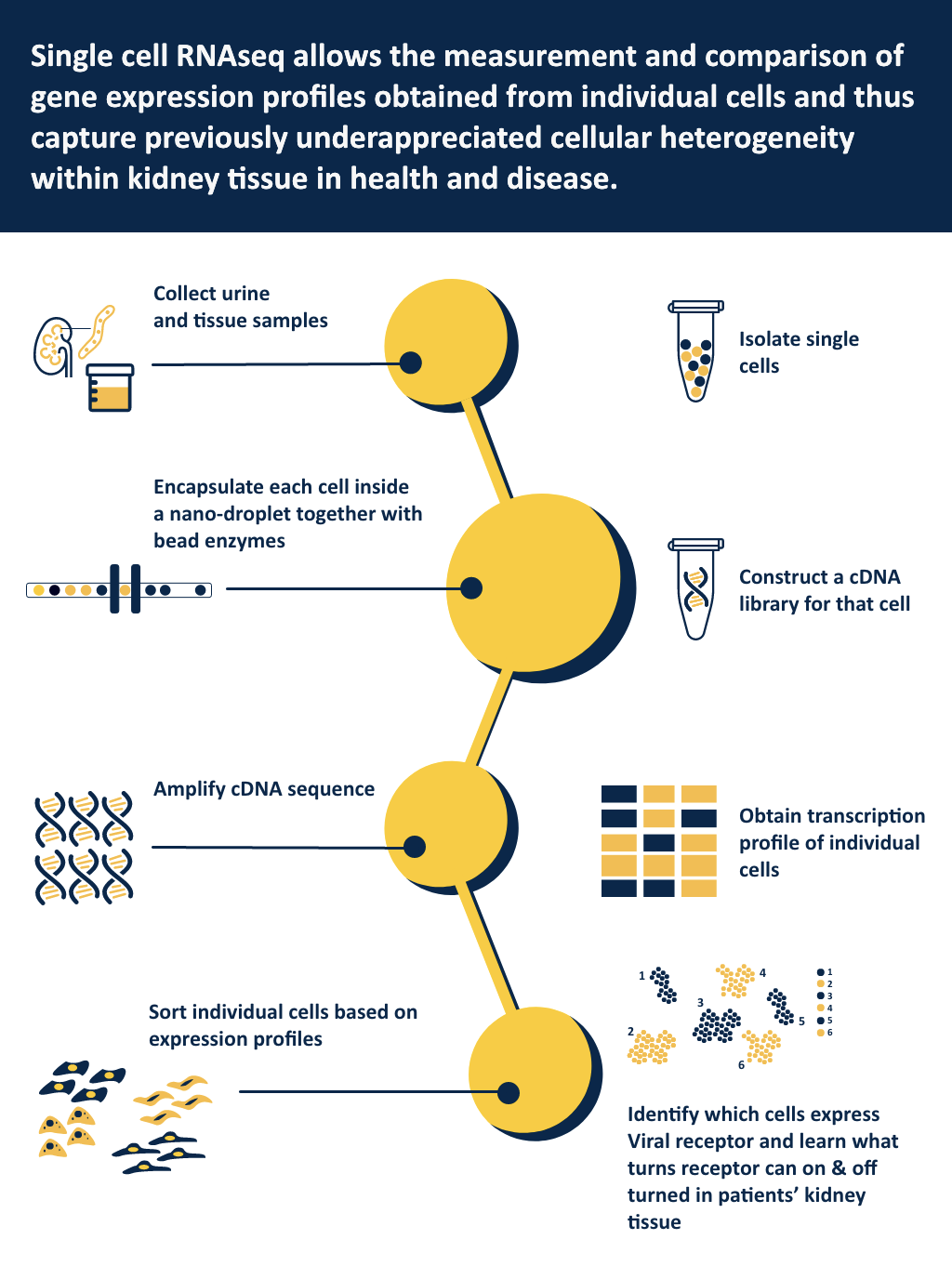
What new piece of knowledge can this new research bring? Coronaviruses need specific keys, cellular receptors, to infect cells in the human body and each organ can teach us how the body response differently to the virus attack. During previous virus outbreaks, like the HIV epidemic, kidneys were a primary target of infection, with devastating complications for survivors. Researchers have just learned in the last months how the SARS-CoV-2 uses cell type specific receptors to infect lung, gut, heart and kidney cells. With this information the Michigan Medicine research team and collaborators around the world set out to learn how these receptors are regulated. The researchers are using a two-pronged approach: In existing data, the team already has in-hand, bioinformaticians learn how the receptors are switched on and off in kidney development and diseases. In parallel, a multi-disciplinary team of clinical nephrologists, molecular biologists, kidney pathologists and computer scientists will study patients with acute kidney injury by COVID19 treated at Michigan Medicine. Using a protocol established just last year, the molecular machinery encoded by cellular RNA can be measured in thousands of individual urinary kidney cells to define cell type specific mechanisms of kidney injury. As urine can be easily obtained, the nephrology team can start this study even during the pandemic with this liquid “biopsy” providing a key window into how the virus damages kidney cells.
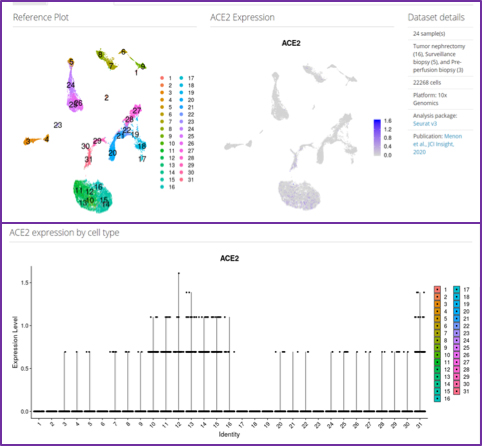
Since 2005, Dr. Kretzler and his team have collected and analyzed data from multiple cohorts of patients with kidney disease in Michigan, all over the USA, and the entire world. Using the available transcriptomics and clinical data from these cohorts, the team has been able to identify specific kidney cells that selectively express SARS-CoV-2 receptors. This now allows identification of key molecular pathways that enable the coronavirus to infect cells, take over their metabolism, replicate and eventually destroy the infected cells.
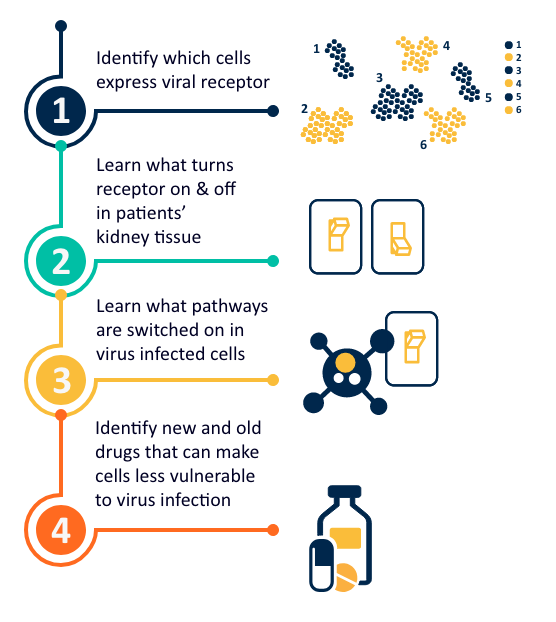
Several therapeutic applications can emerge from this research. The existing data can be used to search for factors that increase virus receptors on cells, leading to more intense infection and subsequent damage in the kidney and other organs. Patients with preexisting conditions such as diabetes, hypertension, autoimmune disease and organ transplantation are very susceptible to coronavirus infections. As the Michigan Medicine team has carefully studied the kidney transcriptomes in these diseases, studies are already underway to learn why the kidneys in these patients are so sensitive to viral damage. The team is also studying how existing medications could change the virus receptors in patients’ cells to make them more or less susceptible to viral damage. These findings can help to develop recommendations of what kind of drugs might make cells more resistant to infections.
The large collections of kidney biopsy data sets coordinated by the Michigan team now allows computer scientists to integrate and study these molecular and clinical data in multi-disciplinary research networks across the US, Europe and China. The research team’s ability to rapidly shift research priorities, addressing the demands and challenges of this unprecedented COVID-19 pandemic, are a testament to the strength of data sharing and scientific collaboration.
The data generated by Dr. Kretzler’s team has benefited from the state-of-the-art University of Michigan core facilities which have rapidly responded to the challenge and are on standby to quickly process patient samples to generate the critical data needed from patients.
The Michigan Team has developed with funding from the NIH a series of data sharing and visualization tools such as Nephroseq (https://www.nephroseq.org/resource/login.html) and Nephrocell (figure 3, http://nephrocell.miktmc.org/), enabling the research community to quickly access data to answer key questions about molecular machinery present in different kidney cells types, how they are regulated in kidney disease and can be targeted for therapeutic attack.