Center co-director, Dr. Mats Lungman’s KLIPP system for CRISPR-Cas 9 featured in informative SciTube video

Enjoy this video which details KLIPP, a “split enzyme” system pioneered by Center for RNA Biomedicine co-director, Mats Ljungman, Ph.D.
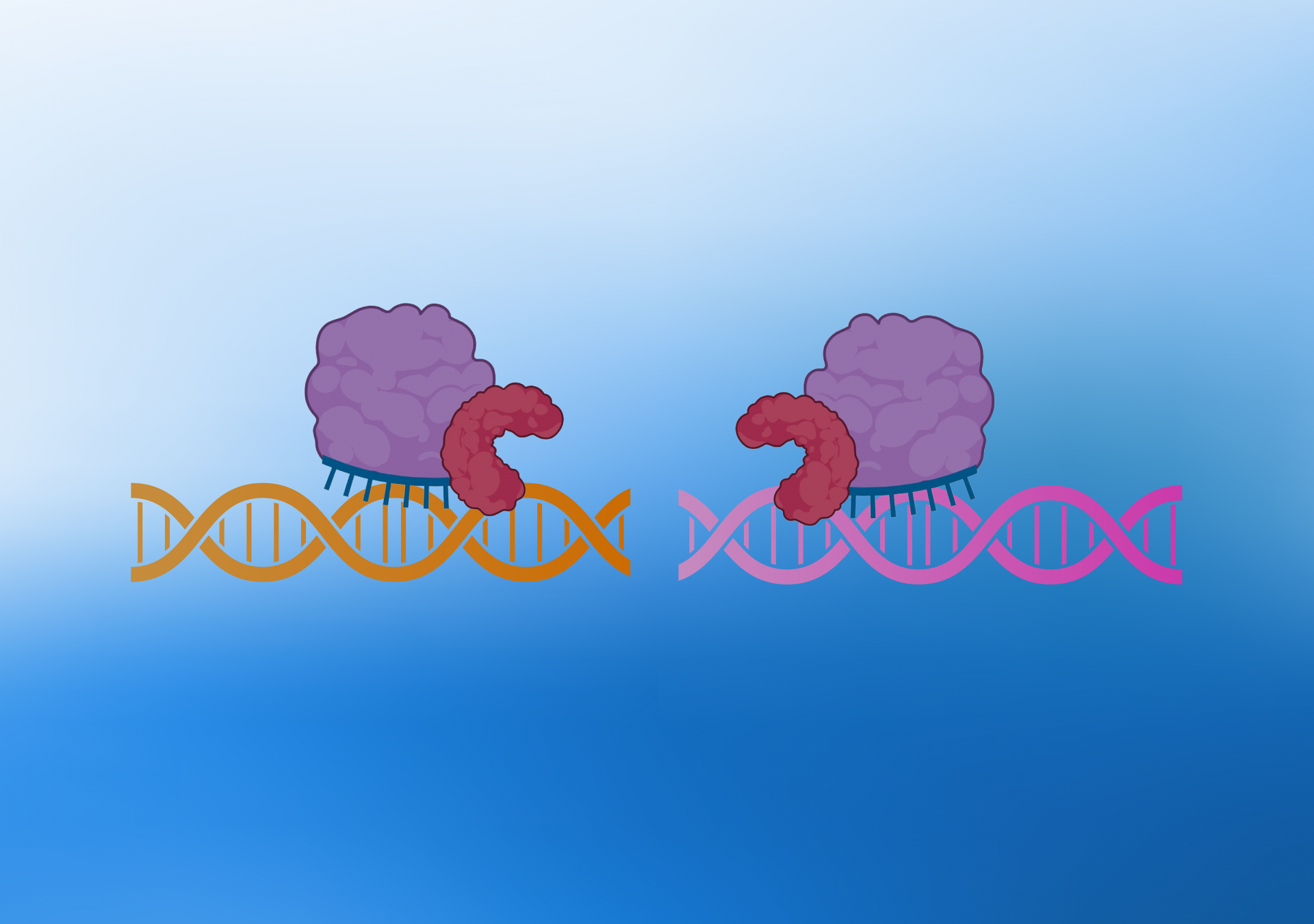
During the KLIPP process, two non-functional Cas9 enzymes fused to Fok1 enzymes are brought together by a pair of guide RNAs. These guide RNAs specifically target sequences near structural variant junctions unique to cancer cells.
In February, the NIH awarded Dr. Ljungman’s lab a $3.7 million grant to continue this groundbreaking work.
Cancer treatments are designed to kill cells within a tumor, but they often affect healthy tissues, leading to serious side-effects. Thus, there is an urgent need to develop new approaches that specifically aim for targets that are unique for tumor cells. A common and early event in carcinogenesis is the formation of chromosomal structural variants. These rearrangements are unique for each tumor and contribute to the growth of the tumor. Structural variants in cancer cells create unique junctions of DNA sequences, which are typically located far apart in normal cells. Thus, these junctions represent a promising new target for precision cancer treatment.
A new approach, called KLIPP, was designed by Dr. Mats Ljungman and his research group at the University of Michigan to specifically target these unique junctions in cancer cells using CRISPR technology.
CRISPR consists of an enzyme called Cas9, which can be precisely targeted to any desired DNA sequence in the genome using a sequence-specific guide RNA. KLIPP consists of a “split enzyme” system, where two non-functional Cas9 enzymes fused to Fok1 enzymes are brought together by a pair of guide RNAs. These guide RNAs specifically target sequences beside structural variant junctions unique to cancer cells.
When two Fok1 enzymes are brought together, they become activated and will cut the chromosomal DNA, promoting cell death specifically in cancer cells. The enzyme complex will also bind to DNA in normal cells, but it will bind far apart because there are no junctions. Thus, no cutting of DNA will occur.
Dr. Ljungman’s research group has obtained strong proof-of-concept for the KLIPP approach, showing induction of DNA breaks specifically in cancer cells. The induction of DNA breaks by the KLIPP treatment leads to toxicity and the demise of cancer cells.
When using the KLIPP approach to target bladder tumors in mice, over half of the mice showed no evidence of tumor tissue remaining after a 4-week treatment period.
When brought into clinical practice, cancerous and normal blood cells would first be collected from a patient and subjected to whole genome sequencing. The sequencing data are then mapped, allowing cancer-specific structural variant junctions to be identified. Targeting pairs of guide RNAs are then designed. The Cas9-Fok1 MRNA and targeting guide RNAs are then encapsulated into lipid nanoparticles. This personalized, precision-targeting medicine is then delivered to the tumor of the patient. KLIPP is a platform therapy that can be used to treat all types of cancers using a single common target.
KLIPP is designed to not cause harm to normal cells. Treatments can be repeated to target different junctions. Therefore, the technology is expected to overcome treatment resistance. The manufacturing of personalized KLIPP medicine is expected to be rapid and cost-effective.