RNA Frontiers: From Mechanisms to Medicine
This year’s symposium explores the dynamic world of RNA, highlighting how fundamental mechanisms and molecular machines are shaping both our understanding of cellular processes and the development of next-generation medical innovations. Through cutting-edge scientific talks and a patient advocacy panel discussion, we will explore a wide range of topics spanning epigenetics, genome editing, RNA structure, and translational research, and discover together how RNA is propelling biological discovery from molecular intricacy to real-world impact in medicine and beyond.
Regular registration is open now through February 20 only!
*PLEASE NOTE: You will be notified that you must have an Iris account in order to register.
- Visit the Registration Page in the link at the button above
- On the sign-in screen, sign in with your Iris account credentials or select “Don’t have an account, Sign Up”
- Follow the instructions to create your account and register for the Symposium
- Creating your account will allow you to log back in to edit your registration responses
- If you experience an issue registering, please clear your cookies and cache (HERE’S HOW) and try to register again. If you are still experiencing an issue, please reach out to Paul Avedisian at paulave@umich.edu.
10th Annual RNA Symposium – RNA Frontiers: From Mechanisms to Medicine
March 6-7, 2026 | Kahn Auditorium, University of Michigan, Ann Arbor Campus
Center for RNA Biomedicine
Join us for a series of dynamic presentations, panel discussions, and collaborative sessions
designed to push the boundaries of RNA research and foster interdisciplinary collaboration.
Dates: March 6 – 7, 2026
Location: Kahn Auditorium and ABC Seminar Rooms
Biomedical Science Research Building (BSRB)
University of Michigan, Ann Arbor Campus
2026 RNA Symposium Speakers

The Center for RNA Biomedicine at the University of Michigan proudly invites you to the 2026 RNA Symposium. Taking place from Friday, March 6 to Saturday, March 7, 2026, this premier event will convene thought leaders and pioneering researchers in the field of RNA science and biomedicine.
Welcome to our 2026 RNA Symposium Sponsors!
Interested in becoming a sponsor for 2026?
Or contact Paul Avedisian at paulave@umich.edu or 734-764-8024
Download the sponsorship flyer pdf here.
Scheduled speakers:

Daniel S. Och University Professor
Director of the Epigenetics Program
University of Pennsylvania
“Epigenetic pathways as targets in human disease”
Abstract:
Chromatin regulatory proteins are frequently mutated or overexpressed in human disease. Because they are enzymes, chromatin proteins are outstanding targets for drug development. Our work focuses on elucidation of epigenetic pathways that might be disease drivers, and epigenetic pathways that might augment clinical treatment.
Our work in cancer focuses on epigenetic pathways utilized by the tumor suppressor p53 and oncogene HIF2alpha; we also study epigenetics of cancer immunotherapy pathways of active and exhausted T cells. In addition, we investigate epigenetics of aging and in regulation of mouse and human memory and diseases of memory, including human Alzheimer’s disease. Aspects of these epigenetic regulatory pathways will be discussed.
Biography
Shelley Berger is the Daniel S. Och University Professor at the University of Pennsylvania and the Director of the Epigenetics Institute at the Perelman School of Medicine. Dr. Berger earned her PhD from the University of Michigan and was a postdoctoral fellow at the Massachusetts Institute of Technology. She previously held the Hilary Koprowski Professorship at the Wistar Institute in Philadelphia. Nationally she received the Ellison Foundation Senior Scholar Award in Aging, the Glenn Foundation Award in Aging, and the HHMI Collaborative Research Award. Dr. Berger is an elected fellow of the American Academy of Arts and Sciences, the National Academy of Medicine, the American Association of Cancer Research, the Academy of Healthspan and Lifespan Research, and the National Academy of Sciences. She has been awarded the Outstanding Investigator Award (R35) from the NIH National Cancer Institute. Dr. Berger’s body of work over twenty-five years helped to launch the “modern era” of chromatin biology and epigenetics. Her early discoveries provided a framework and paradigm for mechanisms of histone and factor-modifying enzymes in gene regulation. Dr. Berger’s research over the last decade uncovered a vital role of epigenetic regulation in mammalian and human health, behavior, and disease, including pioneering discoveries of physiological functions of histone modifications in aging and senescence, cancer, T cell exhaustion, mammalian learning, and memory, as well as revealing a decisive role underlying organismal level behavior and aging in ant societies.

R. Selden Rose Professor of Molecular Biophysics and Biochemistry and Professor of Cell Biology
Director, Yale Center for RNA Science and Medicine
“Co-transcriptional RNA processing yields unexpected versatility in gene regulation”
Abstract:
Many steps in RNA biogenesis occur during synthesis by RNA polymerases. Co-transcriptional activities are commonplace in prokaryotes, where the lack of membrane barriers allows mixing of all gene expression steps. In the past decade, an extraordinary level of coordination between transcription and RNA processing has emerged in eukaryotes. I will discuss our recent work, highlighting the development of RNA sequencing strategies that detect transient transcription and RNA processing intermediates that have led to discoveries of how constitutive and alternative splicing, 3′ end cleavage, and dynamic RNA folding are coordinated during transcription. For example, long read sequencing reveals transcript-specific behavior whereby intron retention across a nascent transcript suppresses 3′ end cleavage, resulting in readthrough transcription. A hallmark of eukaryotic cellular stress responses, I will show that cancer therapy triggers transcriptional readthrough and the production of RNA chimeras comprised of exons from two adjacent genes. These and other examples show that co-transcriptional RNA processing events can impact gene output as dramatically as DNA-based mechanisms, such as transcription factor control.
Biography
I have a BS in Biology from Cornell University and a PhD in Neuroscience from UCSF. I switched gears to RNA biology as a postdoc with Mark Roth, working on the newly discovered SR protein family of splicing regulators. I was inspired to study RNA metabolism in vivo by combining imaging, genomics, and nascent RNA sequencing strategies. I have studied splicing in relation to nuclear speckles and discovered that most introns are removed during their synthesis, or co-transcriptionally. Recently, my lab discovered “all-or-none” RNA processing in which splicing of all introns as well as 3’ end cleavage are coordinated, such that nascent transcripts are either efficiently processed and expressed or inefficiently processed and either degraded or rescued later. My lab has also shown that snRNP assembly occurs in membraneless organelles called Cajal bodies (CBs) and that depletion of the CB scaffolding protein coilin is lethal in zebrafish embryos, due to a deficit in splicing. I was a group leader at the Max Planck Institute in Dresden Germany from 2001-2013. I am currently Professor of Molecular Biophysics and Biochemistry and of Cell Biology at Yale University, where I am also Director of the Yale Center for RNA Science and Biomedicine. I am passionate about climate change, believing that everyone has something to contribute to meet its challenges. I have developed biochemistry curriculum to show the relevance of the discipline to meeting the current and future needs of our planet.

Associate Professor, Biomedical Engineering, Graduate Biomedical Sciences Member, Genetics, Molecular and Cellular Biology, Graduate Biomedical Sciences Member, Neuroscience Program, Tiampo Family Fellowship, Biomedical Engineering, Tufts University
“A splice-switching antisense oligonucleotide approach for pediatric epilepsies”
Abstract:
Variants in ion channel genes are common causes of pediatric epilepsy, often leading to intractable seizures, developmental delay and other comorbidities, which increases risk of death. Pathogenic variants in the SCN8A gene, which encodes a voltage-gated sodium channel critical for action potential generation in the brain, account for ~1% of genetic epilepsies. The voltage sensor in SCN8A domain 1 is encoded by one of two developmentally-regulated mutually exclusive alternative exons, 5N and 5A. We observe that variants in these exons are more likely to cause infantile spasms, a severe seizure type, than variants elsewhere in SCN8A, and that some pathogenic variants affect exon 5 splicing, impacting patient phenotype. Molecular and evolutionary analyses implicate the exon sequences of these and other voltage-gated ion channel alternative exons in splicing regulation. We identified antisense oligonucleotides (ASOs) that shift splicing of SCN8A exon 5N to 5A or vice versa. These ASOs normalize neuronal activity in patient-derived iPSC neurons, and reduce seizures and motor impairment and extend lifespan in a new exon 5N mutant mouse model. Our results demonstrate that splice-switching ASOs can effectively reduce the expression of pathogenic isoforms and rescue both seizure and non-seizure phenotypes. Similar approaches should be applicable to pediatric genetic epilepsies caused by mutations in other ion channel alternative exons.
Biography
Madeleine is the Tiampo Family Associate Professor of Biomedical Engineering at Tufts University, after completing a PhD in Neuroscience from King’s College London on adult neurogenesis and post-doctoral research at MIT on cancer metastasis. Her independent lab focuses on understanding the mechanisms by which the tumor microenvironment contributes to cancer metastasis and resistance to drugs. She has received numerous awards for her research such as a K99/R00 Pathway to Independence Award in 2016 and a DP2 New Innovator Award in 2021 and was voted Exemplary Engineer by the graduate students in her department 3 years in a row for her commitment to promoting diversity, equity and inclusion in biomedical engineering. In 2021, the diagnosis of her daughter Margot with mutations in the SCN8A gene led to her to start research on SCN8A in her own lab, work to develop an ASO for patients with SCN8A, and become an advocate for individuals with epilepsy and other disabilities.

“Prime Assembly with Linear DNA Donors Enables Large Genomic Insertions”
Abstract:
Many diseases are caused by diverse mutations across various loci, necessitating precise therapeutic strategies. However, current genome editing approaches struggle to insert or replace large DNA fragments. This has made it difficult to develop editing approaches that can address most or all mutations among a diverse patient population. As a result, separate editing strategies must often be pursued allele by allele, which is slow, arduous, and expensive.
Targeted insertion of large DNA fragments holds great promise for overcoming these limitations of CRISPR engineering. Strategies employing recombinases or integrases increase the already substantial complexity and delivery challenges associated with prime editing (PE). We have developed Prime Assembly (PA) for the insertion of large DNAs whose ends overlap with the 3’ flaps generated by PE. We used PA to insert DNA fragments up to 11 kilobase pairs. Either single-stranded or double-stranded DNA donors can support PA. PA relies on DNA templates that are easily produced, and it does not require co-delivery of exogenous DNA-dependent DNA polymerases, recombinases, or integrases. PA proceeds in non-cycling cells, suggesting independence from homology-directed repair (HDR) pathways. In summary, PA can initiate “Gibson-like” assembly in mammalian cells to generate gene insertions without dsDNA breaks, recombinases, or HDR.
Biography:
Erik J. Sontheimer, Ph.D., is the Pillar Chair in Biomedical Research and Professor at the University of Massachusetts Chan Medical School, where he is also Vice Chair of the RNA Therapeutics Institute (RTI). He earned his Ph.D. from Yale University in 1992, having completed his thesis in the laboratory of Joan Steitz. He conducted postdoctoral research as a Jane Coffin Childs Fund Fellow with Joe Piccirilli at the University of Chicago. He then joined the faculty at Northwestern University where he continued his work on the roles of RNA molecules in gene expression, including pre-mRNA splicing mechanisms, RNA interference pathways, and CRISPR immune systems in pathogenic bacteria. Among other advances, in 2008 his group reported that CRISPR systems can function via DNA destruction, and they first described CRISPR’s potential for RNA-guided genome engineering. He has received a CAREER Award from the National Science Foundation, a New Investigator Award in the Basic Pharmacological Sciences from the Burroughs Wellcome Fund, a Basil O’Conner Award from the March of Dimes, a Scholar Award from the American Cancer Society, a Distinguished Teaching Award from the Weinberg College of Arts and Sciences at Northwestern, the Nestlé Award from the American Society for Microbiology, the Mid-Career Award from the RNA Society, and election to the American Academy of Microbiology. In 2014 he co-founded Intellia Therapeutics, Inc. for the development of clinical applications of CRISPR gene editing. That same year he also moved to the RTI at UMass Chan Medical School, where he is continuing his research on the uses of RNA molecules in biomedical research and the treatment of human disease. From 2021-2023 he co-chaired the Board of Scientific Counselors at the National Cancer Institute, and he currently serves as a member of the Scientific Advisory Board at Tessera Therapeutics.

Francis S. Collins Collegiate Professor of Chemistry,
Biophysics & Biological Chemistry
Founding Co-Director, Center for RNA Biomedicine
University of Michigan
“Life in Flux: Dynamic RNA:Protein Complex Assembly Shapes Biomolecular Function“
Abstract:
The explosion of cryo-EM structures in recent years has underscored the stepwise assembly of stable biomolecular machines with defined, fixed compositions. In contrast, advances in single-molecule imaging—both in vitro and in live cells—are revealing a very different picture: Many biological complexes are not static but highly dynamic and transient. Instead of persisting as stable entities, their functions emerge from short-lived, fluid assemblies, whose lifetimes and outputs are governed by the kinetics of their components. This chemistry-driven paradigm shift—from rigid machines to kinetically controlled assemblies—offers a powerful framework for understanding gene regulation, proofreading, checkpoint control, and cellular adaptability. This talk will illustrate this evolving view with two case studies: the kinetically programmed exchange behavior of the RNA silencing machinery, and the dynamic assembly of phase-separated RNA-protein structures (RNP granules) in mammalian cells. I will also explore broader implications of this model, including how regulatory signals can fine-tune molecular function by modulating kinetic parameters, rather than altering structure or affinity per se. By reframing molecular cell biology through the lens of kinetic control and spatiotemporal organization, this presentation aims to offer a unifying conceptual foundation across diverse areas of biomolecular science.
Biography
Nils G. Walter is the Francis S. Collins Collegiate Professor of Chemistry, Biophysics & Biological Chemistry at the University of Michigan, Ann Arbor, where he co-directs the Center for RNA Biomedicine and directs the Single Molecule Analysis in Real-Time (SMART) Center. Trained at the Technical University of Darmstadt (Diploma with Hans-Günther Gassen) and the Max-Planck Institute in Göttingen (Dr.-Ing. with Nobel laureate Manfred Eigen), he completed postdoctoral work in RNA biophysics before joining the Michigan faculty in 1999 and rising through the ranks.
Walter’s laboratory develops and applies single-molecule and super-resolution fluorescence methods to reveal how noncoding RNAs and RNA:protein machines fold, interact, and function in vitro and in living cells. His group’s methodological and conceptual advances have illuminated heterogeneous folding and catalysis in ribozymes and riboswitches, the biochemistry of the splicing and RNA silencing machineries, intracellular RNA trafficking and RNA:protein complex phase separation, and new paradigms for ultra-sensitive RNA, DNA and protein biomarker detection. These contributions—published in leading journals including Science, Nature, Cell, Molecular Cell, Nature Nanotechnology and Nature Methods—have established single-molecule fluorescence as a central tool for RNA biology.
A committed institutional builder, Walter founded the SMART Center (NSF MRI support) and co-founded the Center for RNA Biomedicine, initiatives that have expanded campus capabilities in RNA therapeutics and advanced microscopy and achieved a national profile. He also served as Faculty Director of the Biomedical Microscopy Core and as Associate Director of the University of Michigan PREP postbac program, and he has a long record of mentoring successful graduate students and postdocs. Walter’s honors include the Camille Dreyfus Teacher-Scholar Award, the RNA Society Mid-Career Award, election as an AAAS Fellow, and institutional awards for mentoring and service. He has received sustained federal support (including an NIH R35 MIRA) and is author of more than 230 peer-reviewed publications (h-index ≈ 69), reflecting a sustained impact on RNA biophysics, translational RNA science, and microscopy.

T.C. Jenkins Professor of Biophysics
Johns Hopkins University
Talk Title TBD
Abstract: Coming soon!
Biography
Sarah Woodson is the T. C. Jenkins Professor of Biophysics at Johns Hopkins University. She received her PhD in Biophysical Chemistry in 1987 with Donald Crothers at Yale University, and did postdoctoral research in the laboratory of Thomas Cech at the University of Colorado Boulder. Her research group studies how RNA molecules fold, and how RNAs interact with proteins for gene regulation and ribosome assembly. Dr. Woodson was elected a AAAS Fellow, served as the President of the RNA Society and received the Lifetime Service Achievement Award from the RNA Society in 2020 and Ignacio Tinoco Award in Biophysical Chemistry from the Biophysical Society in 2023.
Special guests:

Patient Advocate
Biography
Victoria Gray was the 1st Sickle Cell Anemia patient in the world to be treated with CRISPR gene editing in 2019. After a lifetime of pain, treatments, and hospitalizations for Sickle Cell Disease, she is now symptom-free, working as a patient advocate and international speaker to spread the word about CRISPR and rare diseases to clinicians, scientists, patients, and students. Mrs. Gray is a wife and mother of three children and lives in Forest, Mississippi.
Symposium and Gala Dinner Registration Fees
Symposium Registration Fees*
*All registration fees include the Networking Dinner
| Registration type | Early bird (Oct-Jan 19 ) | Regular (Beginning Jan 20) |
| U-M student/postdoc | $45 | $50 |
| Non U-M student/postdoc | $80 | $92 |
| U-M academic/nonprofit | $100 | $112 |
| Non U-M academic/nonprofit | $160 | $184 |
| Industry/corporate | $350 | $400 |
Networking Dinner
The Networking Dinner is now included in the Registration Fee. No separate registration is needed. All registrants will be automatically signed up for the dinner.
All attendees are invited to join us for a Networking Dinner on the first evening of the Symposium, Friday, March 6, from 5 PM – 8 PM in the Palmer Commons Great Lakes Room
The dinner provides an opportunity for attendees to mingle with speakers, presenters, and fellow attendees in a relaxed and enjoyable setting. Enjoy a memorable evening of connections and inspiration!


Accommodations Information
Room blocks have been reserved at the following local hotels. Please book directly via the links below to make your reservation.
Note: Daily shuttle service may be arranged by guests directly with the hotels located outside of walking distance from the event venue.
Hampton Inn South

Hampton Inn – South
Hampton Inn South Website
925 Victors Way, Ann Arbor, Michigan 48108
734.665.5000
Click the link above to book directly into the Symposium room block. Please note: Symposium room block rate expires Friday, February 23!
The Kensington Hotel

The Kensington Hotel
Kensington Website
3500 South State Street, Ann Arbor, Michigan 48108
866.446.1329
Please note: Symposium room block rate expires Friday, February 13! Click the link above to book directly into the Symposium room block.
The Bell Tower

The Bell Tower*
Bell Tower Website
300 S Thayer St, Ann Arbor, MI 48104
(800) 562-3559
*Short walking distance to conference venues!
Click the link above to book directly on the hotel website.
Inn at Michigan League

Inn at the Michigan League*
Inn at the League Website
911 North University Ave., Ann Arbor, MIchigan 48109
734.764.3177
*Short walking distance to conference venues!
Call to reserve using booking block code RNA26
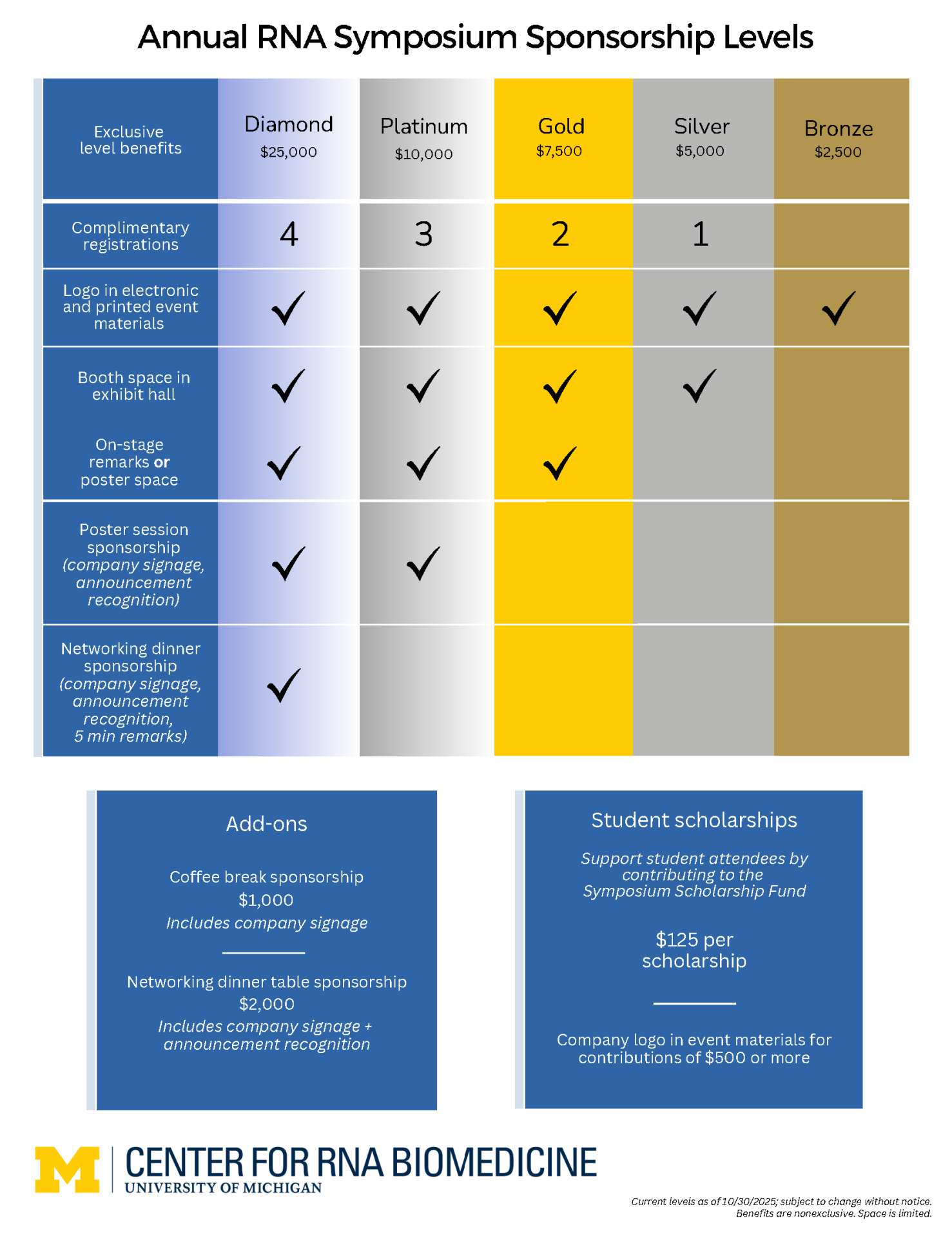
Sponsorship Information
Celebrate a Decade of RNA Innovation!
Join us as a sponsor of the 10th Annual RNA Symposium — a landmark event hosted by the Center for RNA Biomedicine. Gain unparalleled exposure to a dynamic community of trailblazers in RNA science and medicine. Connect and collaborate with leaders in the field, and play an essential role in advancing the next wave of biomedical breakthroughs.
It’s the beginning of an exciting new era in RNA research!
Questions? Contact RNA Center Manager, Paul Avedisian at paulave@umich.edu
Abstract Submission Information
Interested in presenting your work at the Symposium?
Abstract submission date closed January 19, 2026, 11:59 PM EST. Decisions will be made and participants notified after February 16, 2026.
Please Note: A limited number of abstracts may be accepted for the poster session only after the deadline.
We’re currently accepting abstract submissions to be included in the event programming.
Click the link below for more information and to submit your abstract today!
All submissions will be considered for entry into the poster session as well as the opportunity to present on stage. Please indicate in the form below your desire to give a presentation in addition to participation in the poster session. Poster and presentation slots are limited.
A professional panel will review all submissions and select participants based on abstract submission. The panel will be rating each submission based on topic relevance, scientific rigor, and research significance. Please complete the form below to submit your information and abstract.
Submit your abstract by clicking the button below!

Photo: Scott C. Soderberg, Michigan Photography


Click the button below to view a detailed program of the 10th Annual RNA Symposium:
RNA Frontiers: From Mechanisms to Medicine on our Symposium Hub
(Coming Soon!)
Regular registration is open now through February 20 only!
*PLEASE NOTE: You will be notified that you must have an Iris account in order to register.
- Visit the Registration Page in the link at the button above
- On the sign-in screen, sign in with your Iris account credentials or select “Don’t have an account, Sign Up”
- Follow the instructions to create your account and register for the Symposium
- Creating your account will allow you to log back in to edit your registration responses
- If you experience an issue registering, please clear your cookies and cache (HERE’S HOW) and try to register again. If you are still experiencing an issue, please reach out to Paul Avedisian at paulave@umich.edu.